I.INTRODUCTION
Researchers in this paper kept the standardization to the global perspective for analytics to be deployed on cloud and therefore, used Fast Healthcare Interoperability Resource (FHIR) 4.0 HL7 to model data. This standardized endocrine corpus is being analyzed to diagnose diabetes mellitus (DM) and predict comorbidities in this paper. The diagnoses are labeled with ICD-10-CM codes for uniformity and understanding of international medical nomenclature. Other corpus that would be used is ICD-10-CM corpus extracted from the website; www.icd10data.com. ICD-10-CM corpus would be tagged using tagged CSV HL7 standardized endocrine corpus in the textual form. Both these Named Entity Recognition (NER) tagged corpus would give us a medical corpora to be used for unified natural language programming (NLP) techniques for medical context learning for diagnosis, recommendation, prescribing, or treating a particular patient in future.
Previously, Intelligent Medical System prototype (IMP) [1] is proposed for e-coaching and recommendations that uses an incremental learning mechanism. It showed 90% accurate results, and the user satisfaction was 80% having 95% integration with legacy system. It uses the capabilities of interactive dialogue system. Internal knowledge system is built on heterogeneous data acquired from multiple sources validated by knowledge experts by ripple down rules over an interoperable system with adaptable user interface. The patients’ health logs are maintained that generate alerts on observing critical condition of patient for a specific disease. Prototype of IMP was tested in a hospital in Korea. The targeted disease was Thyroid Cancer diagnosis, treated through follow-up and recommendations given by doctors. This prototype was integrated in Hospital Health Management Information System to get 500 patients’ post treatment data that was compliant with FHIR standard. Analytics used decision tree algorithms; Chi-squared Auto Interaction Detection (CHAID) and J48 used majority voting and ensemble learning with 10-folds cross validation to form semantic rules validated by knowledge experts using RDRs. The accuracy with decision trees was 79.79%, J48 gave 80.56% accuracy, and 82.88% accuracy was observed with CHAID. RDRs on other hand were continuously updated with new rules through doctors’ feedback reaching the accuracy level of 90.32%. User satisfaction was evaluated by sharing two questionnaires; user interaction satisfaction was for overall system usage, while user experience questionnaire was to access user interface for emotions and feelings expressed through attitudes after using the system. Clinical Document Architecture complying with electronic medical record (EMRs) was converted to Virtual Medical Records for IMP with 93% accuracy. Next-generation medical systems are seen to have dialogue-driven chat bots with FHIR as mediator standard as depicted by IMP [1].
The rest of the paper is organized as follows. Section 2 refers to the previous propositions in the area of big data analytics for health. Sections 3 and 4 elaborate on significance of NLP techniques in medical domain and how the limitations found are taken care of in this paper by proposing a standardized mechanism for medical context learning keeping to endocrine patients’ data. Section 5 explains the experiments performed to implement the proposed mechanism in section 4. Unified tagging of medical corpora with endocrine patients data and ICD-10-CM corpus for future use by medical community for analysis is presented in section 5. Sections 6 and 7 conclude and elaborate the future direction for experimenting with the common NLP techniques unified for medical context learning.
II.UNIFIED GENERALIZED BIG DATA CLOUD ANALYTICS BUILT ON NLP
S Johnson et al. [2] came up with the unified data model, in year 2000, on which analytics for chronic diseases were built for monitoring purposes with integration of different mining operands. Hadoop Distributed File System lets innovate Big Data Analytics methodologies through altering the replication factor for fault tolerance to maintain data integrity in healthcare databases. The quality of analysis results is directly proportional to the accuracy and completeness of health data. It is also recognized that regional diseases show some unique features known to that region only thus it is said to be phenotypically related.
In healthcare, big data analysis is seen to have the potential for monitoring, prediction, diagnosis, and management of diseases and other systems performing in healthcare at organizational level like insurance and emergency care systems that need speed to respond through in-time recommendations for personalized patient care. The survey from major countries showed that about 72% of medical professionals and students working in different fields were not aware of the term big data and 80% did not witness to see any big data technology been implemented anywhere while 20% said that if implemented it is not effective and is used at a very small scale. Challenges that pose hindrance in adoption of big data technology in health sector are trustworthiness and usability of data, the will to adopt by health professionals, and security concerns. Now, as the awareness is created of smart hospitals, 93% showed interest to use the facility.
Complete healthcare data from different sources forms large data banks that would be accessible through query system to give deeper understanding of medical histories, diagnosis, recommendations, and treatment plans with the underlying risks associated with patients’ profiles. This architecture would enable us to devise a large analytical system set on a uniform data model standardized on FHIR HL7 to analyze heterogeneous data converging in from multiple sources as a major shift in healthcare business models.
III.NATURAL LANGUAGE PROGRAMMING IN HEALTH SECTOR
Textual clinical notes are also a good resource for data extraction for e-Phenotyping, however, challenging due to its free form for which two ways of extraction are there; symbolic and statistical [3]. Symbolic focuses on predefined relations where statistical annotates the corpus of text for finding semantic relationships. There has been growing platforms for NLP processing on text in clinical notes to form interoperable data models, but the first one was MedLEE (Medical Language Extraction and Encoding). Mayo Clinic [3,4], while working on Learning Healthcare System, also devised an NLP pipeline cTAKES (clinical text analysis and knowledge extraction system) that is open source to get clinical rules for symptoms, diagnosis, medication, etc. Research is led to develop a large corpus of clinical text taken from Mayo Clinic in syntactic form. The first machine learning application was applied to Phenotyping in 2007 [5] on cohort of diabetic patients using feature selection via supervised model construction with 47 filtered features ranked on scale for their significance. At that time Naïve Bayes, C4.5, and IBl (Instance Bases Learning algorithms) were used to identify diabetic patients. In another study [6], prescriptions data, ICD-9 coding, and clinical notes from Unified Modeling Language System (UMLS) were employed to come up with Phenotyping model using SVM rheumatoid arthritis. This study took all features structures and unstructured to show that SVM as in [7–9] was as good on unrefined feature sets as was on engineered. Noise in data could not be ignored for which Halpern et al. [10] used framework of Agarwal et al. [11] XPRESS (extraction of phenotypes from records using silver standard) to build platform for extraction of features and building models. These researchers assumed that large data sets would mitigate the effect of label errors by setting bounds and would generate results as good as in small data that is clearly labeled (Gold Standard). Phenotyping was defined as three pillars: (i) complex relationship between multiple features, (ii) it is understandable by medical knowledge domain experts, and (iii) its definition is transferable into new domain knowledge. Researchers used this definition to introduce high-throughput phenotyping [12] that was unsupervised transformed in a scalable format. These phenotypes were clustered in correspondence with the diseases and validated by medical experts. PheKnow cloud tool by Henderson et al. [13] evaluated phenotypes derived from previous medical literature and associates them to the biomedical standard codes; latest International Classification of Diseases (ICD) codes, SNOMED-CT (Systemized Nomenclature of Medical – Clinical Terms), or MeSH, etc. and ranks as per relativity thus limiting the need of medical expert review. Automated Feature Extraction for Phenotyping (AFEP) extracted features from medical resources like Wikipedia and Medscape, to list UMLS concepts to train classifier. Feature sets are more refined using NLP and ICD codes are given to develop hybrid applications like ElasticNet on Logistic Regression Model. Surrogate-Assisted Feature Extraction extended AFEP to include other resources like Merck Manuals, Mayo Clinic UMLS, and Medline plus Encyclopedia removing noise from phenotypes to classify manually labeled patients on gold standard.
Long Short-Term Memory (LSTM) is known for automatic concept tagging in clinical notes as referred to in [14]. Clinical word embedding model is trained to learn context of problem, tests, and treatments given to patients stated in practitioners’ notes. Clinical NER is quite challenging when the labeled corpus is not available. Manual feature engineering has always remained troublesome before the annotated clinical corpus was given access to. Tokenized feature engineering is also considered limited in its use as was proposed earlier in conditional random fields (CRF). Deep learning is adopted in NLP-related tasks as we experienced in fast.ai [15,16]. In CRF, LSTM RNN model promised to derive contextual features for clinical concept extraction but had limitations to understand complex word semantics. Other good versions for contextual word embedding were seen as Embeddings from Language Models (ELMo) adopted features of language model to perform better than Glove and LSTM, etc. but lacked understanding of clinical domain. In recent research [14], bidirectional LSTM-CRF model embedded in ELMo was compared with other state-of-the-art models for performance in clinical context. Ensemble of ELMo and LSTM-CRF best performed with the improved performance of 3.4% in terms of F1-score. These recent breakthroughs in embedding approaches using LSTM, ELMo and Bidirectional Encoder Representations from Transformers (BERT), etc. still need a common standard mechanism for integrating clinical context learning [17].
In [18], researchers have found that similar problem of classifying multiple labels, in four data sets with as large as having 0.5 million labels, is solved through eXtreme Multi-label text Classification (XMC). The deep pretrained transformer NLP models gave state-of-the-art results for sentence classification and albeit label sets [19]. Application of these deep pretrained transformer models in XMC problem at first did not give good results for data sparsity and huge volume. Therefore, X-Transformer was proposed to solve XMC problem by fine-tuning the transformers with a scalable approach. The pairs of (instance, label) had to be formed and the scalable function needed training such that the pairs with high relevance had high score. This hypothesis was challenging to solve. The first challenge was to determine model parameters and GPU memory size when XLNet was applied to XMC model. There was a difference in MNLI data set in GLUE and the 1 million labels in XMC data set so the classifiers size had to differ for both the data sets making XMC problem harder to solve. The other issue was sparsity of data. In XMC data set, only 2% had more than 100 instances to train. 98% of data had long tailed labels with very few instances. This sparsity issue had not been resolved before for a large data set as in XMC. There were two methods to solve this challenging XMC problem; (i) fine-tune deep transformers or (ii) the transfer learning that has been discussed earlier in many contexts of word2vec, GPT, or ELMo having bidirectional LSTM model trained on large data set. The option of transfer learning, however, economical was set aside as it limited the model capacity for adaptation. Deep learning approaches were found better than TF-IDF in extracting neural semantic embeddings in text. XML-CNN contains single-dimension convolutional neural nets to sequence words embeddings to label text. Recent breakthrough in AttentionXML having Bi-LSTMs and learned labels through scoring mechanism gets the models trained with hierarchical label trees. AttentionXML also takes care of negative sampling of labels to avoid back-propagating the bottleneck classifier layer. NLP community is seeing the shift to pretrained and fine-tuned models to achieve GLUE benchmark levels, NER and question answering chat bots, etc. XLNet and RoBERTa are the latest advancements in pretrained transformer models. XLNet, BERT, and RoBERTa are taken to fine-tune and give X-Transformer to solve hypothesis of high scored function for instance relevant to the label. BERT and RoBERTa are same in speed, while XLNet is slower. In performance, RoBERTa and XLNet were better, and there was not much significant difference in the results.
Researchers in this paper have explored Bi-LSTM and keras on TensorFlow framework for text mining and classification for diagnosis using proposed unified NER tagging mechanism to be built on ICD-10-CM Tagged corpus (Fig. 1). Researchers intend to explore X-Transformer and other recent models [20,21] proposed for XMC problem to find solution to medical context learning elaborated in the proposed mechanism in section 4.
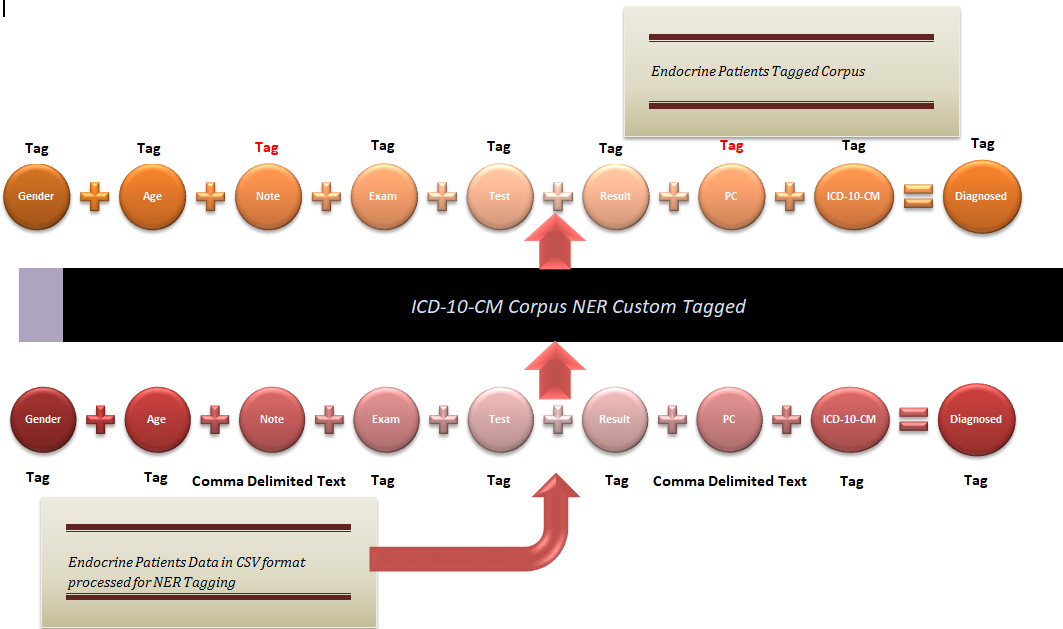 FIGURE 1. Proposed mechanism for custom unified NER tagging of ICD-10-CM corpus tagging for diagnosing endocrine diseases.
FIGURE 1. Proposed mechanism for custom unified NER tagging of ICD-10-CM corpus tagging for diagnosing endocrine diseases.
IV.PROPOSED DIAGNOSTIC NER TAGGING OF CORPUS FOR DM AND ITS COMORBIDITIES
Wu et al. [22] had worked on automated free text mining to find phenotypes in medical context. NLP process implementation or transfer has always remained difficult on new data or settings. Paper [22] proposed distributed representation mechanism to train and reuse NLP models through identified phenotype embeddings in patient profiles. Twenty-three phenotypes were extracted from 17 million documents of anonymous medical records from South London Maudsley, NHS Trust in UK, for application on 6 morbidities. The experiments were done to reevaluate NLP models for identification of 4 phenotypes. Proposed approach selects the best NLP model using two measures for quantification of reductions in duplicate and imbalance wastes. The proposed approach also guides in validation and retraining of NLP models to perform up to 93% to 97% accuracy. Adaptation and reuse of NLP models with phenotype embeddings in new settings are more effective with higher efficiency than blindfolded adaptation of NLP models.
In this paper, researchers tried to propose the standard mechanism for clinical context learning that was missing in [17]. The authors of this paper have devised a big data diagnostic analytics model for DM and its comorbidity diseases. This analytics needs uniform clinical context learning for its HL7 FHIR v4 standardized diagnostic data model to be analyzed in interoperable and generalized settings over the cloud. The phenotype fields contained in our custom endocrine patients’ corpora in.csv format would be used to tag the corresponding values in NER format associated with the diagnosis of DM and its comorbidities.
The NER tagged corpora of endocrine patients’ diagnostic profiles would then be used to tag ICD-10-CM corpus generated from (www.icd10data.com). Iterating the same step, this ICD-10-CM corpus would be used to tag comma separated “Note” and “PC” fields in our custom endocrine patients’ corpus as shown in Fig. 1.
The analytical techniques used in our research are applied on the custom corpus of 14407 DM patients with co-existing diseases with 33185 instances mapped onto unified standard data model set on FHIR HL7 schema labeled with ICD-10-CM codes.
V.EXPERIMENTATION WITH NLP TECHNIQUES
In our experimentation, Gradient Paperspace GPU data science template frameworks are explored for reusing NLP models mentioned in [22] for medical context learning.
Experiment 1: Transformers + NLP template is used to create our python notebook. The CSV corpus of 14407 DM patients’ data set is tagged with comma delimited “Note” and “Diagnosed” fields to train and predict “Diagnosed” disease labels using NLP Sequential NER model made on LSTM, Keras, and TensorFlow. (Note, Diagnosed) tagged values are paired as an albeit form for patients as in [18] to find the right sequence to diagnose the corresponding disease. Figure 2 shows a dry run of the code that was tried with LSTM and Bidirectional LSTM both that gave near to zero percent accuracy which led us to other experiments with other entities like “Exam” or “Test” fields which do not hold sparse values as the “Note” field in our data set.
 FIGURE 2. The implementation of TensorFlow.Keras Sequential() model with LSTM to tag the predicted diagnosed diseases through clinical “Notes” having near to zero accuracy.
FIGURE 2. The implementation of TensorFlow.Keras Sequential() model with LSTM to tag the predicted diagnosed diseases through clinical “Notes” having near to zero accuracy.
Experiment 2: The same Sequential() model having TensorFlow.Keras and Bi-LSTM was applied to the other set of label values (“Exam,” “Disease”), in sequence of (instance tag, label tag) pair for f(x, y) to predict the most probable disease for the prescribed examination by the doctor as per patient’s problem. In previous experiment “Note” field had sparse values in the form of long sentences and therefore model accuracy was near zero.
“Exam” field used as tag in place of “Note” had much smaller instances to tag corresponding “True” disease as “Diagnosed” (Fig. 3). RMSProp trained model gave accuracy up to 0.5631 where model validation accuracy was 0.8846 in only first 2 epochs (Fig. 4). RMSProp optimizer was found fast learner in comparison to Adam in Bi-LSTM Keras Sequential() model. It took less than 10 epochs to train the model with RMSProp where Adam was hard to train even with 50 epochs in default settings (Fig. 5).
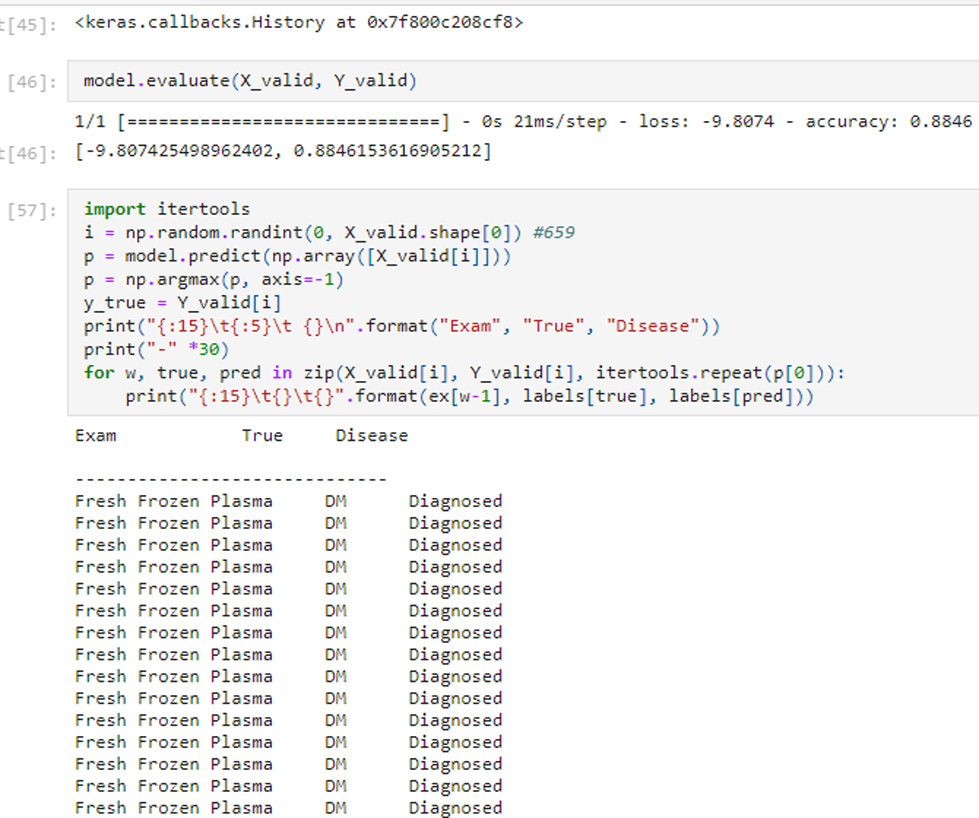 FIGURE 3. The (‘Exam’, ‘Disease’) pair on reaching maximum validation accuracy of 0.8846 tags corresponding Disease Label as ‘Diagnosed’.
FIGURE 3. The (‘Exam’, ‘Disease’) pair on reaching maximum validation accuracy of 0.8846 tags corresponding Disease Label as ‘Diagnosed’.
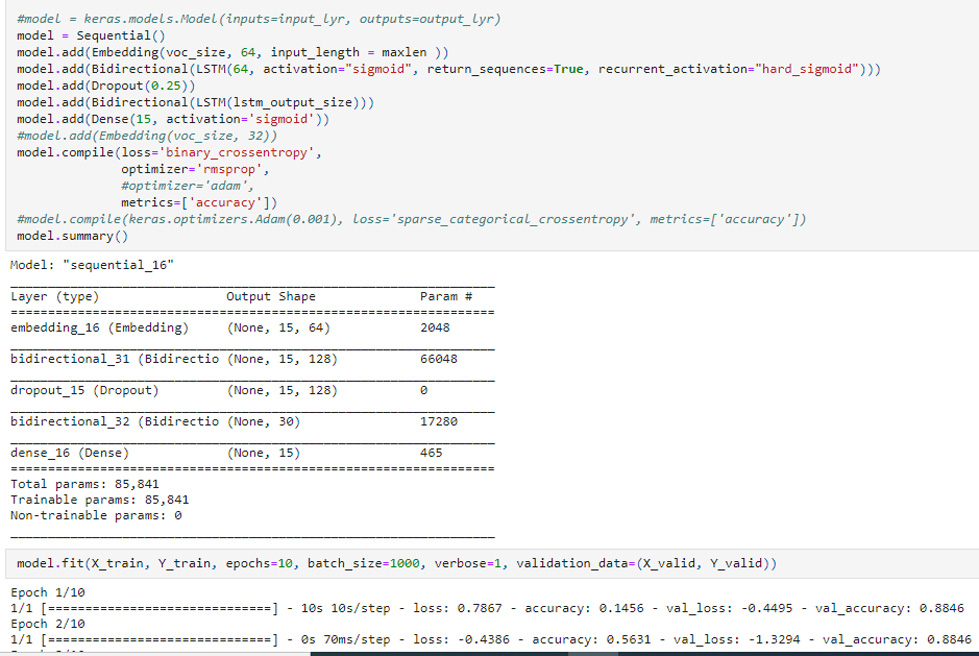 FIGURE 4. Keras Bi-LSTM Sequential() model trained on RMSProp optimizer giving validation accuracy of 0.8846 in first 2 epochs.
FIGURE 4. Keras Bi-LSTM Sequential() model trained on RMSProp optimizer giving validation accuracy of 0.8846 in first 2 epochs.
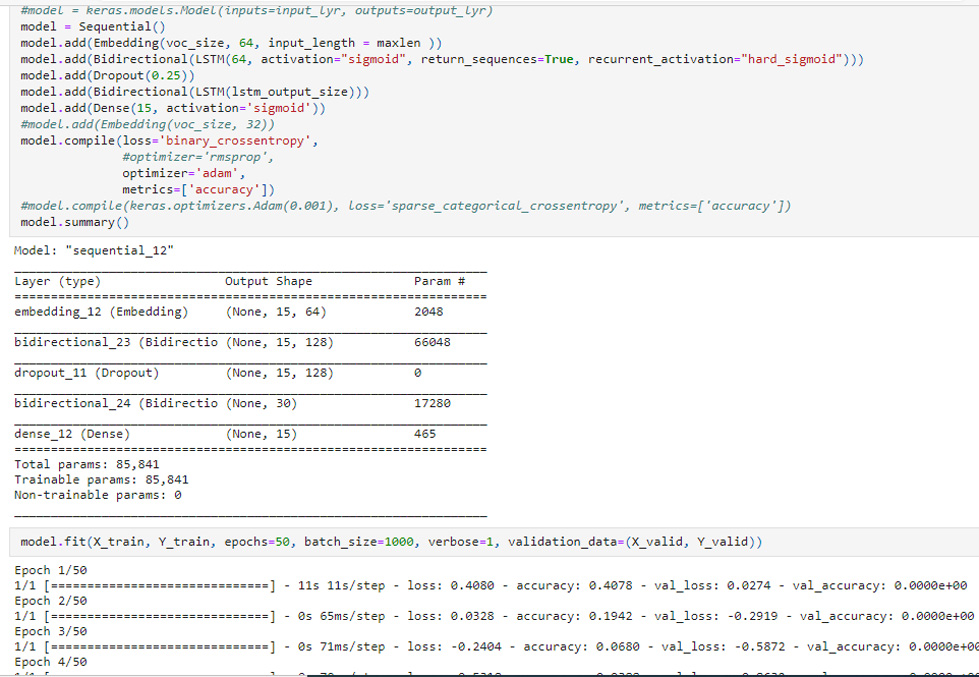 FIGURE 5. Keras Bi-LSTM Sequential() model trained on Adam optimizer giving validation accuracy of 0.
FIGURE 5. Keras Bi-LSTM Sequential() model trained on Adam optimizer giving validation accuracy of 0.
VI.CONCLUSION
The authors of this paper mainly proposed the mechanism (Fig. 1) in section 4 for NER tagging of unified medical corpora for standardized medical context learning. Few known NLP libraries like Keras over TensorFlow framework were reused for medical domain learning on given corpora of endocrine patients and ICD-10-CM coding scheme. LSTM and Bi-LSTM were both tested and validated for Sequential() model and achieved relatively good accuracy of 0.8846 with single parameter “Exam” to predict True disease tagged as “Diagnosed” (Fig. 3).
VII.FUTURE WORK EXPLAINED
Known NLP methodologies used in fast.ai as mentioned in previous section or others like, NLTK, BERT, LSTM, Keras, etc., would be experimented to implement our proposed mechanism. The mechanism as demonstrated in Fig. 1 is for NER tagging of endocrine patients data set and ICD-10-CM corpus for uniform learning of medical context. These experiments are being conducted at lower level with access to the NLP frameworks and libraries in open-source cloud platforms; Gradient Paperspace, etc., for high-performance computing. Researchers also intend to explore and implement XMC [18] to find solution to medical or clinical context learning [17] elaborated in the proposed mechanism in section 4 with higher accuracy.