I.INTRODUCTION
With the popularization of medical imaging technology and equipment, more and more ultrasound, magnetic resonance imaging (MRI), computed tomography (CT), and other imaging methods are available for medical diagnosis. These diagnostic technologies are increasingly and widely used in clinical research and treatment plans in daily life [1]. In the diagnostic analysis of medical images, medical image segmentation usually forms an important step [2], such as renal medical image segmentation, retinal OCT image segmentation [3,4], and abdominal CT scan segmentation [5,6]. Although clinically experienced professional doctors may provide very accurate segmentation, it is often costly and labor-intensive in terms of standard clinical settings. By contrast, automatic lesion segmentation of medical images can greatly reduce labor and increase efficiency, so automatic medical image segmentation technology [7] is demanding in clinical diagnosis and scientific research. Nevertheless, the current medical image segmentation technology has poor long-term context dependence and cannot retain more spatial features, so the segmentation accuracy is not high, which leads to great possibility of misdiagnosis, high missed diagnosis rate, and difficult advancement of automatic diagnosis.
CNN-based UNet [8] model is extensively used in modern medical image segmentation. By replacing pooling operation with resampling, UNet helps to restore fine-grained information of the target object, but the feature map constantly gets smaller during convolution, resulting in fewer spatial features. Pooling operation [9] reduces the feature vector output by the convolutional layer and improves the result at the same time. Nonetheless, it has limitations due to long-term dependence relationship [10,11], cannot easily adapt to individual difference changes in size, shape, and texture [7], nor is it able to extract more spatial features. Transformer (ViTs) [12] solves the problem of long-term dependency, but is inferior to CNN and RNN in local feature capture. TransUNet [5] combines the advantages of both transformer and UNet, but it will create massive computing requirements for high-resolution images and cannot maintain local continuity around the face, resulting in failure to retain more spatial features. DS TransUNet [8] establishes long-term dependence between different scale features, which also effectively integrates multiscale feature representation from encoder to improve computing efficiency. However, it cannot extract higher and more global spatial features. In order to extract more spatial features, we put forward a U-shaped network with PST-UNet network framework (positive distribution data Swin transformer), which establishes a long-term dependency relationship between different scale features. While integrating multiscale feature representation from encoder more effectively, it preserves spatial feature information to the maximum extent to improve the segmentation accuracy.
In order to enhance the accuracy of kidney medical image segmentation and retain more spatial features, a PST-UNet network model was developed. This model yields superior segmentation accuracy in medical CT image segmentation. The rest of the paper is organized as follows. Section II presents the RELATED WORKS. Section III introduces THE PROPOSED METHOD. Section IV introduces the EXPERIMENTS AND DISCUSSION. Section V presents CONCLUSIONS.
II.RELATED WORKS
Based on application of CNN network in medical image segmentation, Neerav Karani et al. [13] designed a segmented CNN to connect two subnetworks and test time adaptability as a way to solve the robustness problem. [1] BANet also has encoder and decoder network structure like UNet, but it uses pyramid edge feature extraction module to collect two-dimensional image edge information, which coordinates with cross-feature integration module CFF to achieve information complementation between features at different levels. MUNet [14] is a medical image segmentation framework based on feature pyramid to enable accurate segmentation of medical image. [15] Shi Tianyi fused 2DUnet and directional field models with 3DUnet models to form a lesion segmentation method featuring multimodel fusion. Medical image segmentation architecture DDU-Net [16] has two encoders and a decoder, which is applicable to skip connection of dual encoder network. The above-mentioned CNN-based image segmentation models [17] have been successfully applied to solve various problems in computer vision by using encoder and decoder framework for image segmentation tasks. Nonetheless, the selection of threshold value and regional division criteria is greatly affected by image intensity or texture information, and there is insufficient ability in global context connection, so limited spatial features are retained and segmentation accuracy is restricted.
In terms of transformer application in medical image segmentation, Swin transformer [8] introduced sliding window mechanism and achieved great success in downstream tasks. ConvNeXt [2] of pure convolutional neural network model was redesigned based on standard ResNet to replace Swin transformer. By using an improved medical image segmentation program, ConvNeXt has significantly reduced the number of parameters. TransUNet [5] established a self-attention mechanism from sequence to sequence and a hybrid CNN-transformer architecture was used to employ detailed high-resolution spatial information from CNN features and the global context encoded by transformers. Then, upsampling was performed on the self-attention features encoded by transformers, and accurate positioning was achieved by combining different high-resolution CNN features skipped from the coding path. In the medical image segmentation model TransUNet+ [8], a feature enhancement module was designed to strengthen skip connection for fusion of multiscale features, but edge feature information was still insufficient. TransUNet+ follows the U-shaped structure of TransUNet, which also contains the CNN encoder, transformer encoder, and CNN decoder. Skip connection was redesigned using column vectors of the score matrix to enhance skip characteristics. DS TransUNet [7] was established based on dual-scale encoding mechanism, which used a dual-scale encoder based on hierarchical Swin converter to learn multiscale features. Each medical image is segmented into nonoverlapping blocks at large scale and small scale, respectively. With the two blocks of different scales as input, the proposed dual-scale encoder subnetwork can effectively extract coarse-grained feature representations of different semantic scales. In the same way as the traditional U-shaped architecture, the extracted context features will be upsampled by the designed decoder and fused with the unified feature representations from the encoder by skip connections. The network based on TransUNet not only possesses ability of CNN to extract local features but also enjoys the advantage of transformer in remote context capture, which makes up for the CNN defects. At the same time, the accuracy of medical image segmentation is enhanced through the self-attention mechanism. Although the existing TransUNet series models can retain spatial features, retained spatial features are insufficient, so CT renal medicine images have a low accuracy in lesion segmentation.
In order to retain more spatial features and improve the lesion segmentation accuracy of renal medical images, the PST-UNet network model was designed to enable better accuracy in medical CT image segmentation.
III.THE PROPOSED METHOD
In this paper, the PST-UNet architecture model will be introduced in detail, as shown in Fig. 1. The PST-UNet network has a UNet structure composed of FB module, PSTP module, PSTU module, and SSM module. The FB (dual-scale fusion block) module fuses the feature information integrate and recognize more spatial features. PSTP module performs downsampling and linear transformation of the information derived from PST as a preparation for the stacking of small Swin transformer blocks in the next layer. Through hierarchical method, PST uses Swin transformer to introduce GELU [18] according to the characteristics that the segmentation data of renal lesion tend to be normally distributed in this project, so that more features of different scales can be captured. The PSTU module inputs the upsampled feature information to the PST module to retain more spatial feature information. SSM samples the image information and simultaneously sends the image information and sampled feature information to the output end, so as to retain more spatial features and achieve better segmentation accuracy of renal lesion. The whole PST-UNet network constitutes the UNet structure, in which the encoder part has PSTP module, encoded information as the FB input, and the FB output is used as the decoder input, while decoder is composed of PSTU module and SSM module.
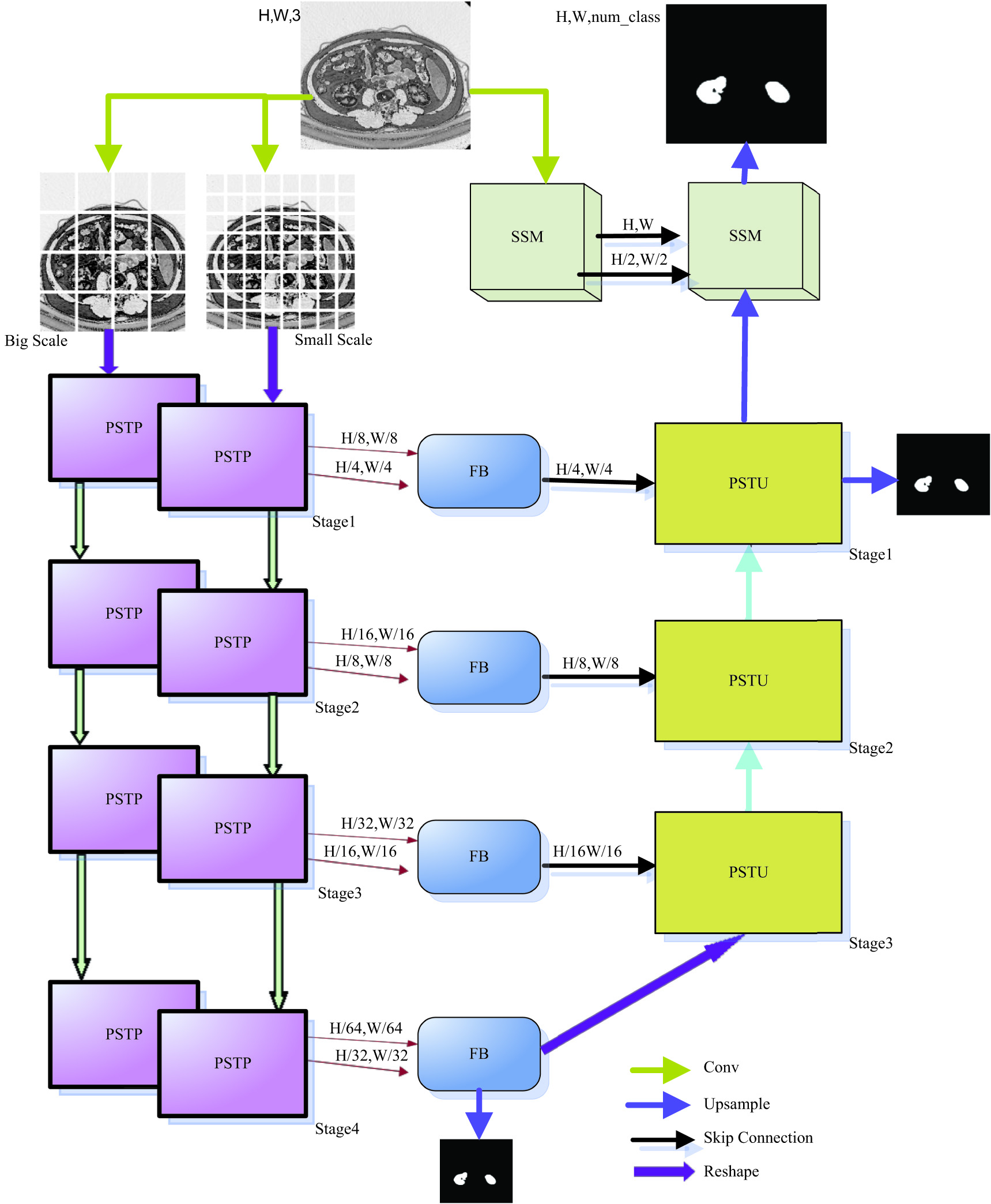 Fig. 1. PST-UNet system model.
Fig. 1. PST-UNet system model.
A.PSTP MODULE
As the core part of self-attention module transformer [8], it allows global dependence and context extraction of renal lesion features, and it uses global attention of the aggregate sequence to reconstruct each token of the sequence. The self-attention mechanism constructs query vector, key vector, and value vector based on input vector. The three matrices are ∈ , ∈ , and ∈ . The formula for constructing these three vectors is as follows:
is an element in the input sequence, and the matrix can be calculated in the same way: Then, based on these three vectors, one input is reconstructed into the new vector : where Q, K, and V are query vector, key vector, and value vector, respectively; is the embedding dimension, and is a query.The combined influence of all input sequences on each location, i.e., self-attention value, is calculated as
The self-attention mechanism establishes a global dependence relationship and has a larger receptive field than CNN, which strengthens the receptive field and accesses more context information. However, the self-attention mechanism is a result of filtering out unimportant information and screening out important information, which has inferior effective information extraction capability than CNN. In order to extract more spatial effective information and improve the precision of renal lesion segmentation, PSTP module was proposed.
As shown in Fig. 3, PSTP boasts PST module function and patch downsampling function. PSTP adopts two-scale encoder mechanisms to extract different scale features. First, a given R3 × H × W image is converted into a patch embedded sequence with two scales of R(H/4 × W/4) × C and R(H/8 × W/8) × C. Then, as the input of two parallel PSTPS, a PST module performs rough and fine parallel encoder of the two scales information. After coding, the two kinds of information are segmented into patches and downsampled to reduce the resolution and increase the depth dimension. Meanwhile, in order to collect more valuable multiscale spatial integration features, the two kinds of downsampled feature information are connected to the input end of FB module for subsequent processing. Parallel PSTP modules have four stages for rough feature and fine feature parallel encoder, as shown in Fig. 2.
 Fig. 2. PSTP module and swing transformer blocks.
Fig. 2. PSTP module and swing transformer blocks.
The PST module consists of small Swin transformer block and full GELU activation function, as shown in Fig. 3. Swin transformer block and full GELU activation function are used in the four stages of encoder: stage1∼stage4.
For the lesion data of renal medical images, it is preferably to extract as much continuous feature information as possible. Nonetheless, some piecewise linear functions such as ReLU are unsmooth, and some breakpoints are not differentiable. For instance, zero point is nondifferentiable. When processing data, the mean value of neural network is usually required to be 0. As the activation function of renal lesion data, ReLU will affect the network segmentation accuracy.
The above suggests that, considering the normal distribution characteristics of renal medical image data in this project, GELU activation function should be selected. That is, Swin transformer is used in every stage of encoder, then ReLU activation function is replaced completely and GELU activation function is used in all. Two scales are adopted at the same time. Coarse-grained features can be located at large scale, while finer grained features can be captured at smaller scale. In this way, it is possible to extract more comprehensive spatial feature information, enhance the model robustness, and improve the segmentation accuracy. First, the medical image was segmented by convolution operation into n (H/n,W/n) patches and mapped into C-dimension by linear embedding. Each patch did not need extra position information, which was input into the PST module of each stage as patch tokens (small block images composed of multiple pixels by linear mapping) for Swin transformer block and full GELU activation function processing. In the encoder part, when the characteristic information goes through stage1∼stage3, the number of PST-transformed tokens gradually decreases, with dimension gradually increased. Then, 2 × 2 adjacent feature patches of each group are patch merged and spliced together for 2× resolution downsampling. The data of tokens are reduced by an exponential multiple of 2. The image dimensions are increased by the same exponential times of 2 at the same time. At stage1∼stage4, the image resolution output is H/n × W/n, H/2n × W/2n, H/4n × W/4n, H/8n × W/8n, respectively, with corresponding dimensions being C, 2C, 4C, and 8C, respectively.
B.FB MODULE
In order to retain more spatial features, semantics should be integrated and fused. Thus, the fusion block FB (dual-scale fusion block) is proposed [7].
The FB module employs a multihead attention mechanism and utilizes a standard transformer to generate tokens of a specified size based on the feature map of each branch. These tokens are subsequently combined with the token sequence generated by another branch, and self-attention is performed. At each stage, the module only carries out two single-layer self-attention operations, enabling more nonlinear transformations and enhancing the network’s feature learning capability. The module is capable of integrating and fusing features of two scales obtained.
C.PSTU MODULE
The PSTU module is proposed to retain more spatial features and improve the segmentation accuracy of renal lesions, as shown in Fig. 3.
The decoder end has three PSTU modules, each of which includes a Swin transformer block, a full GELU activation function, upsampling, and jumper wire from the FB. In stage 4, the encoder outputs PSTU after FB processing as the initial input at the decoder side. In every stage, the decoder increases the upsampling resolution of the feature map by 2 times and reduces the output dimension by 2 times. Therefore, the output resolutions of these three stages are (H/16) × (W/16), (H/8) × (W/8), (H/4) × (W/4), respectively, and the dimensions are C, 2C, and 4C, respectively. In PSTU, the input feature information is first upsampled twice and then connected with the features mapped by jumper wire at the stage corresponding to encoder. Then, the aggregated features are output to Swin transformer to model the remote dependence of Swin converter, and GELU activation function is used for all. GELU maximally saves the spatial features for data with normal distribution characteristics, and the data of renal lesion features normal distribution. Hence, more long-term dependent features can be extracted during the upsampling to acquire more comprehensive global context information, so that more spatial feature information can be retained more effectively to improve the lesion segmentation accuracy in renal medical imaging.
D.SSM MODULE
In order to better retain spatial features and improve the segmentation accuracy of renal lesions, the SSM module was proposed, which consists of the left-half SSM and the right-half SSM, as shown in Fig. 2. The original image information is input into the left-half SSM of the module and then the left-half SSM performs 3 × 3 convolution of the original image features. By downsampling via the GELU layer, more local resolution features of (H/2) × (W/2) are obtained. In the meantime, the PSTU output of the decoder in the first stage is used as the input of the right-half SSM, and the feature information after downsampling of the left-half SSM is also sent to the right-half SSM through the jumper wire. The right-half SSM fuses the two input features, implements 3 × 3 convolution, and performs upsampling through the GELU layer. At the same time, through connection with another jumper wire, the left-half SSM sends the original image feature information to the right-half SSM for feature fusion at different scales, so that spatial features of the image are retained more accurately. Finally, the upper and lower sampling features are connected with the underlying feature mapping of the original image to output the original image.
E.LOSS FUNCTION
To measure image similarity, the Dice coefficient is utilized, and the images are augmented and trained using simple data augmentation techniques. The first method is , which represents the weighted intersection over union IOU loss. It is used to calculate and optimize the model during iterations. The second method is , which represents the weighted binary cross-entropy loss function. This loss function is also effective in determining the similarity between the predicted image and the label. During the training phase of this project, the loss function comprises both and . The most sensitive outputs to the training results are the encoder’s fourth stage, , and the decoder’s first stage, , as shown in Figure S1. Therefore, the total loss function of this training phase is
The hyperparameters α, β, δ, and γ are empirical values. Specifically, it is recommended to set α and β to 0.2, while δ and γ should be set to 0.2 and 0.6, respectively.IV.EXPERIMENTS AND DISCUSSION
In this chapter, we conducted a comparison between our proposed lesion segmentation method that utilizes kidney medical imaging data and the state-of-the-art DS-TransUNet approach. Furthermore, we carried out ablation studies on the PST-UNet network to examine the influence of various network modules on the accuracy of the segmentation results.
A.DATASETS
KiTS2019 is a competition item for the MICCAI19. We need to train 2D dataset, read 2D slices from 3D CT body data to segment renal tumor lesions. From 2010 to 2018, patients with one or more kidney tumors at the University of Minnesota Medical Center were candidates for the database, and the KiTS2019 dataset was labeled with 2 categories: kidney and kidney tumors. We used 45424.png CT kidney datasets. Each png image had a file resolution of 256 × 256 × 3, with 7995 data for training datasets, 1090 data for validation datasets, and 36339 data for test datasets.
B.IMPLEMENTATION DETAILS
PST-UNet networks of two scales were used, with one encoder based on swin_base_patch4_window7_224 and the other encoder based on swin_tiny_patch4_window7_224, both of which were pretrained and initialized. The model was trained with an SGD optimizer, with the learning rate initialized to 0.01, the momentum at 0.9, and the weight decayed to 1e-4. PyTorch and PyCharm were used to establish all the network models, and the models were trained on A100-SXM4-80GB GPU. All the network models were trained for 1000 rounds, but cosine annealing and early stop were set in the experiment to adjust the learning rate, and multiscale methods were used for training of all experiments. Simple data enhancement was also carried out by random rotation, vertical flip, and horizontal flip.
In this experiment, Dice, the intersection over union mIOU, precision, recall of the two sets of label values, and predicted values were used as the main evaluation indexes to measure the similarity between predicted value and real label value.
C.COMPARISON WITH EXISTING METHODS
Experiments were carried out on the tumor lesion segmentation dataset of renal medical imaging to verify the segmentation accuracy of DS-TransUNet against the dataset. DS-TransUNet adopted double scale, one patch of which was (4,4) and the other patch was (8,8). The combination of two branches (4,8) and double-scale encoder mechanism were experimentally studied. In addition, comparison was made with other different U network architectures, such as 3D U-Net [19] with network structure similar to standard U-Net [20]. Instead of using simple convolution, residual 3D U-Net in encoder [20] and preactivated residual blocks 3D UNet [20] and 2D Unet [21] were used. In addition, 3D UNet [21] of 3D full convolutional neural network based on UNet architecture with the results shown in Table I.
Table I. Comparison of the segmentation effect of existing UNet networks on renal tumor lesions
| Framework | Year | Dice |
|---|---|---|
| DS-TransUNet | 2023 | 0.91276 |
| 3D U-Net [ | 2019 | 0.8504 |
| Residual 3D U-Net [ | 2019 | 0.8573 |
| Preact. Res. 3D U-Net [ | 2019 | 0.8513 |
| 2D Unet [ | 2020 | 0.6775 |
| 3D UNet [ | 2020 | 0.7823 |
In Table I, 3D U-Net variants have relatively low segmentation accuracy, DS-TransUNet network has optimal segmentation effect on renal lesions, with a segmentation accuracy of 0.91276, which is obviously superior to the previous UNet network. It suggests that Swin transformer has greater potential than the previous UNet.
D.ABLATION STUDY
In order to correctly evaluate the ability of each module in the proposed PST-UNet network framework in segmenting lesions in renal medical imaging, an ablation study was performed on the segmentation of renal lesions. All the ablation experiments adopted dual-scale encoders, including two scales of [128,256,512,1024] and [96,192,384,768]. The main modules used in the ablation study include
- 1)Instead of using our ablation module, only the DS-TransUNet network was used to check the dice, precision, recall, and mIOU values of the network in terms of segmentation of renal lesions. The experimental results are shown in Table II:
- 2)Introduction of full GELU
| Framework | Year | Dice | Precision | Recall | mIOU |
|---|---|---|---|---|---|
| DS-TransUNet | 2023 | 0.91276 | 0.93693 | 0.80861 | 0.77584 |
| PST-UNet + GELU(ours) | 2023 | 0.92350 | 0.93499 | 0.89028 | 0.78109 |
| 2023 |
The full GELU activation function was used in the PST-UNet network framework model. At encoder and decoder ends, GELU activation function was used instead in standard 2-dimensional convolution to improve the ability of extracting continuous spatial features. The experimental results are shown in Table II.
- 3)Introduction of full GELU and use of FB module
Full GELU was introduced into PST-UNet model, and joint convolution FB module was used for fusion at different scales to extract as much continuous spatial feature information as possible. Then, the extracted multiscale continuous spatial features were fused to achieve the optimal segmentation accuracy of renal lesions. The experimental results are shown in Table II:
According to Table II:
- 1)When only the DS-TransUNet network was used, the accuracy rate, precision rate, recall rate, and average intersection over union of renal lesion segmentation were 0.91276, 0.93693, 0.80861, and 0.77584, respectively, which were relatively low compared to the use of our ablation module.
- 2)When PST-UNet network used full GELU activation function, the accuracy rate, the precision rate, recall rate, and average intersection over union of lesion segmentation in renal medical imaging were 92350, 0.93499, 0.89028, and 0.78109, respectively. Compared with renal lesion segmentation using DS-TransUNet, the accuracy rate was increased by 0.01074, the precision rate was decreased by 0.00194, the recall rate was increased by 0.08167, and the average intersection over union was increased by 0.00525. This indicated that the precision of renal lesion segmentation was superior to DS-TransUNe network when only GELU module was introduced into the proposed PST-UNet network.
- 3)The accuracy rate of lesion segmentation in renal medical imaging was 0.92381 when both GELU and FB were used in PST-UNet network, which was 0.01105 higher compared to the use of DS-TransUNet, respectively. This suggested that renal lesion segmentation had optimal precision when GELU and FB were used in the proposed PST-UNet network, as shown in Fig. 4.
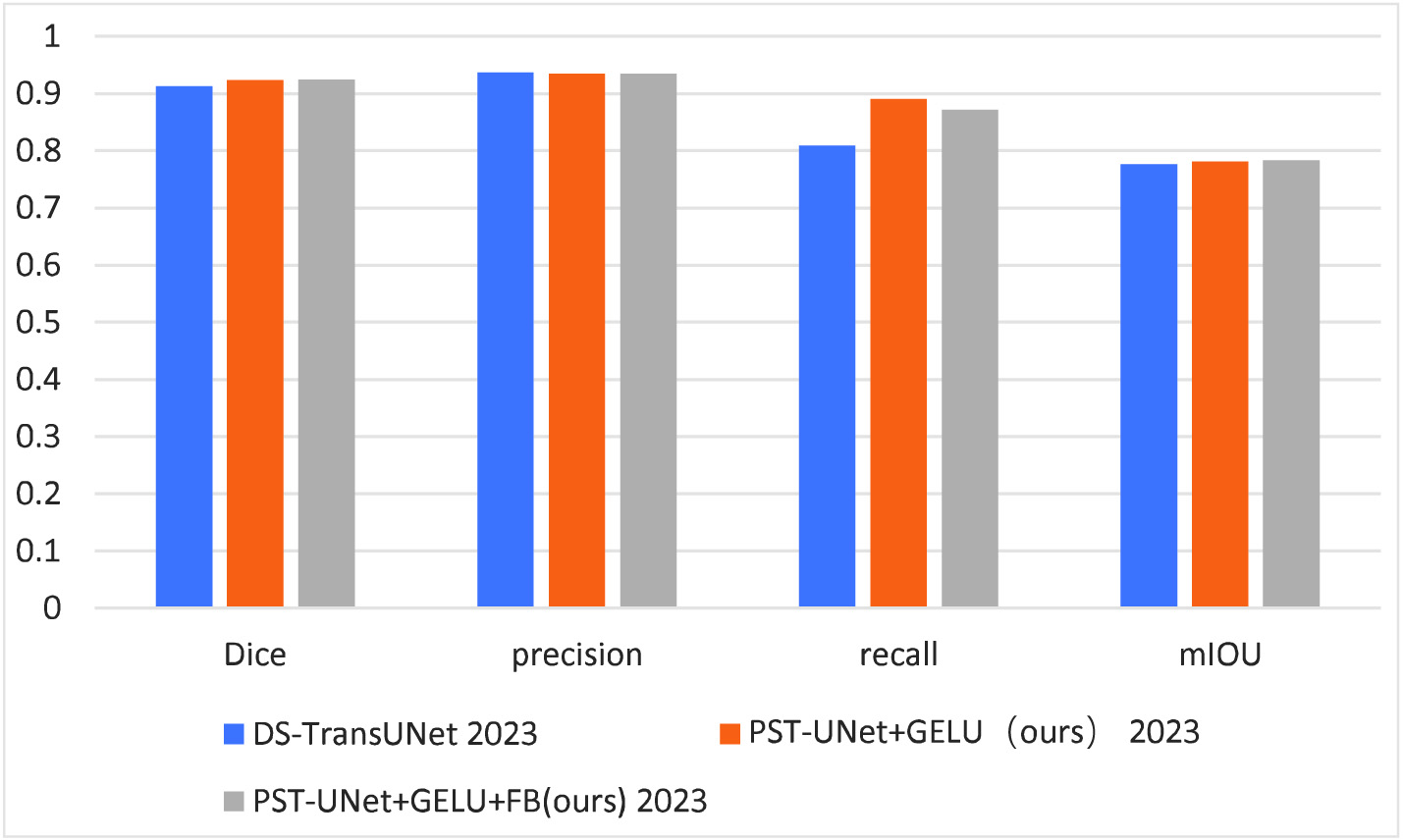 Fig. 4. Comparison of modules used for ablation experiment.
Fig. 4. Comparison of modules used for ablation experiment.
In visualizations with reference to different ablation modules, the test results of renal lesion data were compared, as shown in Fig. 5.
It can be found that undersegmentation occur in the segmentation of renal lesions by DS-TransUNet, PST-UNet + GELU, which some lesions were not detected. PST-UNet + GELU + FB had stronger global context encoding and semantic differentiation, better noise inhibition effect, with fewer false positives in the test.
V.CONCLUSIONS
The paper proposed two types of windows and Swin transformer [8,24] and a parallel encoder mechanism PST-UNet network model, which were used in the lesion segmentation of medical images. This network model reserved more spatial features and enhanced the segmentation accuracy. By introducing GELU + FB module, it became possible to extract more continuous, multiscale, and advanced spatial fusion features, with the lesion segmentation accuracy higher compared to the existing methods, which was expected to improve the level of smart medicine.