I.INTRODUCTION
Semantic segmentation is a technique to segment different objects in an image from a pixel perspective and to annotate each pixel in the original image. It is one of the most basic techniques of computer vision processing and has a wide range of applications such as automatic driving, unmanned aerial vehicle (UAV) navigation [1], remote sensing images [2], medical diagnosis [3], etc. Among them, medical image segmentation is an important topic in the medical image processing community [4].
Magnetic Resonance Imaging (MRI) is based on the fading time of the magnetic field of hydrogen atoms recorded by the magnetic field induction coil after the external magnetic force is withdrawn. MRI is mostly utilized in diagnosing and treating brain tumor disease because of its excellent ability to provide high-resolution views of soft tissue anatomy. To obtain comprehensive information for accurate segmentation, multimodal MRI scans with different imaging parameters are usually required in brain tumor segmentation. Commonly used modalities include fluid-attenuated inversion recovery (FLAIR), T1-weighted (T1), contrast-enhanced T1-weighted (T1ce), and T2-weighted (T2). Different imaging techniques are suitable for different situations. Images of different modalities capture different pathological information and they can effectively complement each other [5,6], which plays a key role in segmenting multiple types of brain tumor regions such as edema (ED), necrotic non-enhancing tumors (NCR/NET), and enhancing tumors (ET). Therefore, the detection of brain tumors in MRI images using semantic segmentation methods has become the focus of many studies [7].
Deep learning-based semantic segmentation methods have become the mainstream of medical image processing, and many semantic segmentation methods have been studied and proposed. Fully Convolutional Networks (FCN) [8] is the pioneer of deep learning in semantic segmentation, proving that neural networks can be trained for end-to-end semantic segmentation of images, and now it becomes the most popular method. U-Net method [8] proposes an “encoder-decoder” architecture based on FCN, which uses deconvolution in up-sampling to increase the number of feature maps while reducing the number of feature maps. By adding a jump connection between the corresponding layers of encoding and decoding to preserve the low-level features of the image. The SegNet method [9] also uses an “encoder-decoder” architecture with maximum pooling of nonlinear upsampling and then convolution of sparse upsampling maps to improve the resolution of segmentation. However, the CNN-based method mentioned above all have problems such as low accuracy and insensitivity to details limited by the receptive field of convolutions, leading to difficulty in characterizing the global dependencies of features.
To alleviate this problem, researchers have employed Recurrent Neural Networks (RNN) to model inter-pixel dependencies, enabling sequential processing of pixels to establish global contextual relationships and improve segmentation. Transformer-based models have been introduced to the study of brain tumor segmentation and receive great attention in the field of computer vision. Dosovitskiy [10]. Proposed the Vision Transformer (ViT) model, applying the Transformer model from the text domain to the image domain. The Transformer-based models exhibit a better ability to capture global contextual information by employing a self-focused mechanism, but there is still the problem of the high computational cost of the change method. And then Swin Transformer [11] introduces a network architecture with a sliding window and hierarchical design, which reduces the computation while considering the acquisition of local features and achieves advanced performance in semantic segmentation. However, there also exists problems of low locality inductive bias in the Swin transformer model, which means that large amounts of training data are required to achieve satisfactory visual performance.
In this paper, we propose a method of multimodal information fusion segmentation for brain tumor segmentation, aimed at achieving fuller utilization of multimodal information for accurate segmentation in MRI. Specifically, the segmentation framework proposed in this paper can be divided into a feature extraction backbone network, a semantic information processing sub-network, a feature reasoning module, and an upsampling network. The feature extraction backbone network adopts the ResNet50 architecture for preliminary feature extraction to extract deep features and multiple shallow features. Moreover, a Semantic Information Processing Module (SIPM) is introduced to achieve free Multiscale feature enhancement and extraction, the core of which is to build a new SI-EE (Semantic Information Enhancement and Extraction) module. Then, a novel Multimodal Feature Reasoning Module (MFRM) is constructed to process both the backbone network feature information layer and semantic feature information layer, and the features of different modalities can be further interactively fused to obtain finer lesion segmentation results.
In summary, the main contribution of this paper can be concluded as follows:
- 1.First and foremost, we proposed a method of multimodal information fusion segmentation to better implement brain tumor segmentation, and a feature extraction backbone network, a semantic information processing sub-network, a feature reasoning module, and an upsampling network are included. The different modules built above result in the following two contributions.
- 2.To effectively extract the deep features and multiple shallow features, the SIPM is designed to achieve free multiscale feature enhancement and extraction by fusing the information of the two feature layers with a new Semantic Information Enhancement and Extraction (SI-EE) module.
- 3.Both the semantic feature information layer and backbone network feature information layer can be obtained with the help of SIPM, and a novelMFRM is introduced to further interactively fuse the obtained features, which can lead to a refined lesion segmentation result.
II.RELATED WORKS
Image Semantic Segmentation: In recent years, with the rapid development of deep learning technology and its wide application in various fields, image semantic segmentation based on deep learning (ISSDL) has also received a lot of attention from researchers across the world [12–16]. And ISSDL can be divided into two major categories [17–19]: ISS based on the regional (ISSR) and ISS based on the pixel (ISSP).
For the ISSR techniques, a regional convolutional neural network (regions with CNN features, RCNN) was proposed [20,21], which fuses candidate regions generated by the selective search (SS) algorithm on the basis of visual features extracted by CNN to achieve a multi-task objective for target detection and semantic segmentation. SPPNet [22,23] network was introduced, which inserts a spatial pyramid pooling player [24] (SPP player) after the RCNN convolutional layer to reduce the repetitive computation process in feature extraction. But these methods still have the limitations of generating too many candidate regions, irregular shape of regions, and a large amount of network operations.
For the ISSP technique, Shelhamer [8] designed the classical fully convolutional network (FCN), which performs pixel-level classification of images in fully supervised learning, and lots of methods based on FCN are proposed [25–27]. Chen [28–30] proposed a DeepLab series improvement scheme, DeepLab network introduced fully connected conditional random field (FCCRF), structured prediction of the coarse segmentation map after FCN, and did image smoothing operation to achieve edge optimization, used convolution with holes to expand the perceptual field of the feature map, and finally output a complete semantic segmentation result of the image. and further optimized to DeepLab-V2 and DeepLab-V3.
Feature Encoding: The encoder-decoder architecture approach has also been proposed to address the balance between performance and efficiency of semantic segmentation, as well as the high computational complexity and memory consumption problems posed on high-resolution feature maps. Ronneberger et al. [13] proposed the classical U-Net network for implementing semantic segmentation of biomedical images, which uses a downsampling operation to gradually reduce the resolution of the feature map in the encoding phase, and a downsampling operation to gradually restore image detail and resolution in the decoding phase. The U-net++ was further developed as a more powerful architecture for medical image segmentation. Badrinarayanan et al. [9,31] proposed a SegNet-Basic network for solving image semantic segmentation tasks in the fields of autonomous driving and intelligent robotics to achieve end-to-end pixel-level image segmentation. And then SegNet was applied to automated brain tumor segmentation for four MRI modalities by Alqazzaz et al. [32]. Noh et al. [33] propose a fully symmetric DeconvNet network based on VGG16, which alleviates the limitations of existing methods based on fully convolutional networks. And DeconvNet was also utilized for Simultaneous Localization and Mapping (SLAM) to improve the drift in long-run odometry [31].
Feature Fusion: Feature fusion is studied to solve the problems of excessive computation, long training time, and severe memory consumption. Liu et al. proposed a technique to add global context to a fully convolutional network for semantic segmentation, and performed many critical tasks [34,35]. Their method uses the average features of the layers to add features at each location.
Ghiasi [36] designed the Laplacian pyramid reconstruction and refinement model (LRR) using the Laplacian pyramid algorithm for the reconstruction of shallow features. Attention mechanism [37–39] with a soft weighting of multiscale features at each pixel location and proposed an advanced semantic image segmentation model that is jointly trained with multiscale input images and an attention model. RefineNet [40] was first proposed by Lin [41], a generalized multi-path refinement network for high-resolution semantic segmentation, which utilizes all available information from the downsampling process to perform high-resolution prediction using remote residual connections. And lightweight RefineNet was further developed for effective and efficient semantic image segmentation [42]. Zhu [43] proposed a brain tumor segmentation method based on the fusion of deep semantics and edge information in multimodal MRI, achieving a more sufficient utilization of multimodal information for accurate segmentation by introducing a semantic segmentation module, an edge detection module, and a feature fusion module.
III.METHODS
A.OVERALL STRUCTURE
The overall framework of multimodal information fusion segmentation proposed in this paper for brain tumor segmentation is shown in Fig. 1. It can be divided into two parts: encoder and decoder. With more detail, the process can be divided into a feature extraction backbone network, a semantic information processing sub-network, a feature reasoning module, and an upsampling network. The feature extraction backbone network adopts the ResNet50 architecture for preliminary feature extraction to extract deep features and multiple shallow features, and then send these features to SIPM to achieve free Multi-scale feature enhancement and extraction. SIPM first fuses the information of the two feature layers, the core of which is to build a new SI-EE (Semantic Information Enhancement and Extraction) module, because the information obtained after fusion contains a lot of semantics information. Thus this module mainly enhances and then extracts semantic information to better realize the segmentation task. Through the first two parts, a backbone network feature information layer and a semantic feature information layer can be obtained, and then through the constructed MFRM (Multimodal Feature Reasoning Module) to process it, its main task is to further interactively fuse the obtained features of different modalities to obtain a more refined lesion segmentation result, and finally obtain the segmentation result through the upsampling network.
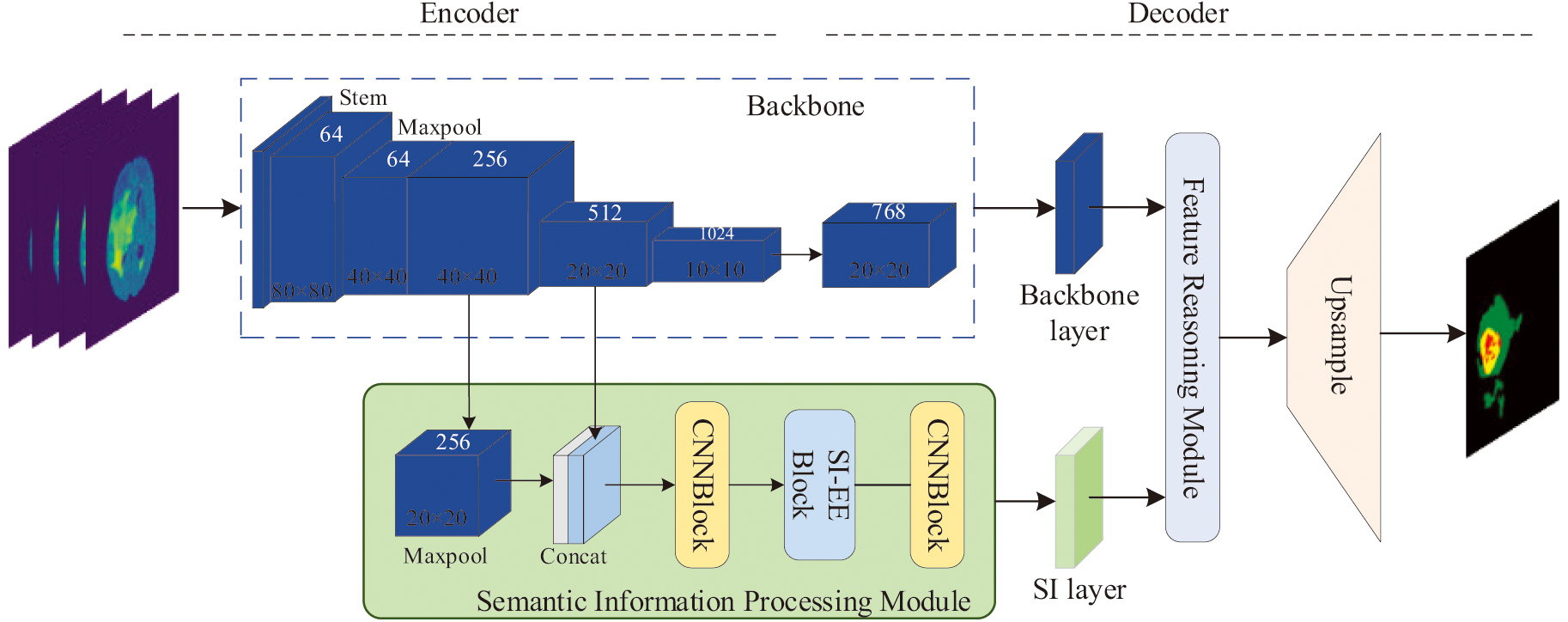 Fig. 1. An illustration of the method proposed in this paper and two parts are included: the encoding part and the decoding part.
Fig. 1. An illustration of the method proposed in this paper and two parts are included: the encoding part and the decoding part.
B.SEMANTIC INFORMATION PROCESSING MODULE
SIPM is to process semantic information, the core of which is the SI-EE (Semantic Information Enhancement and Extraction) module proposed in this paper. Its structure is shown in Fig. 2. The task of this module is mainly to target a large amout of the semantic information of the method is enhanced first, and then extracted, so that the obtained feature information is more detailed and more suitable for subsequent segmentation tasks. Semantic feature information reflects a global feature of homogeneous phenomena in the image and describes the slowly changing or periodically changing surface organization structure and arrangement rules in the image. However, the quality and contrast of the low-level information extracted by the ResNet backbone network (such as pixel values or local area attributes) are usually low, and images captured in dark environments usually have poor visualization quality, resulting in unclear texture details, which cannot be well acquired. And use this low-level information. This paper exploits the statistical properties of textures, which focus on the distribution analysis of images, such as intensity histograms. The first part enhances the texture details of low-level features by histogram equalization after the enhancement step, making it more detailed and suitable for segmentation.
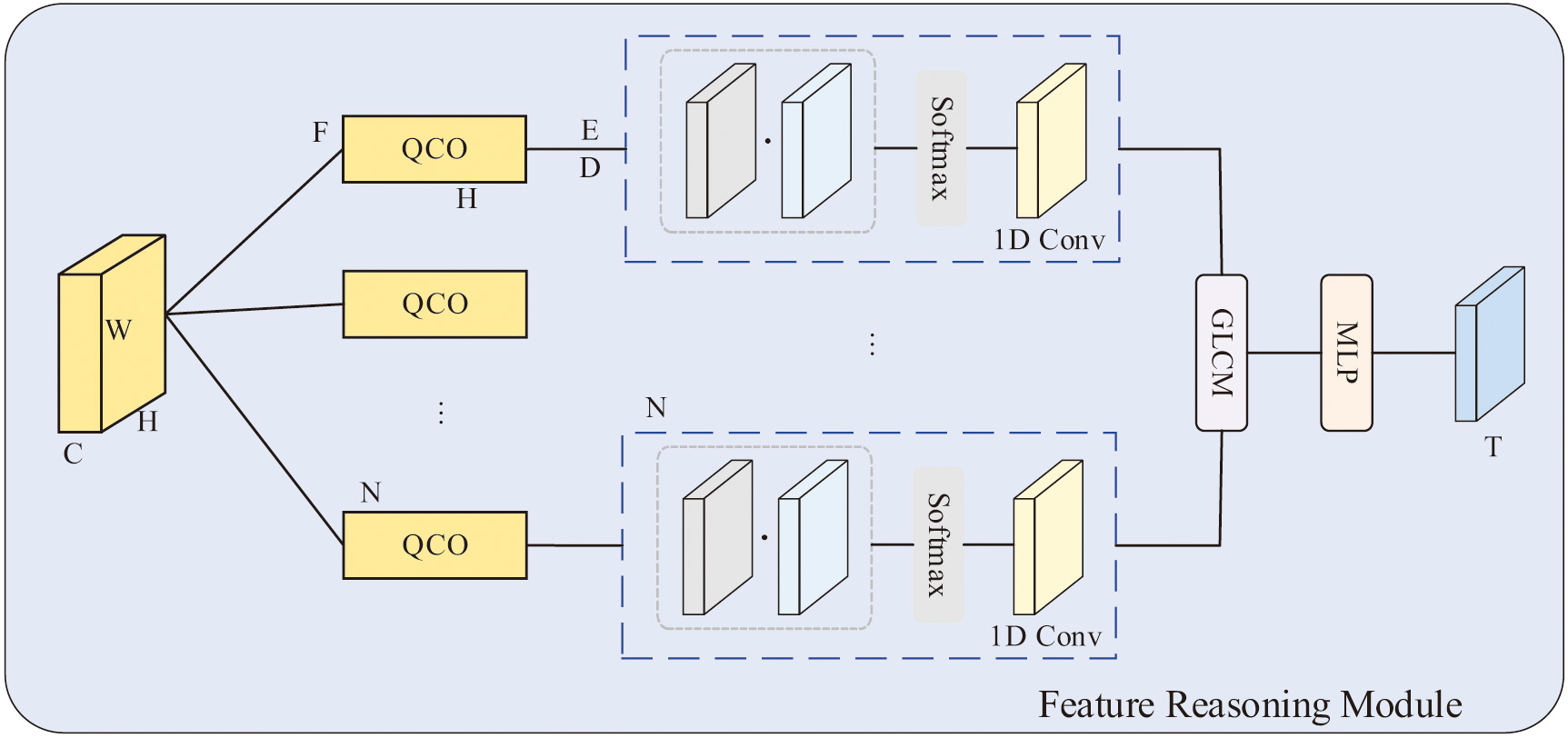 Fig. 2. Semantic information enhancement and extraction module.
Fig. 2. Semantic information enhancement and extraction module.
First, in the generated histogram, its horizontal axis and vertical axis represent gray level and count value respectively, which are expressed as feature vectors H and F respectively. For the histogram quantization, the statistical information of the count value F is reconstructed, and the reconstructed is . Each corresponding level is also converted by the formula :
where N represents the total number of gray levels.Then we introduce the concept of quantization and counting operators to describe the intensity of semantic information. Input the feature map, quantized coding map, and statistical features obtained by QCO, where D plays the role of the histogram. And get the new quantization level from the original quantization level L via D:
where , and denote three different 1×1 convolutions and a Softmax performed on the first dimension acts as a non-linear normalization function.We then update each node by fusing features from all other nodes, resulting in the reconstructed quantization level
Afterward, the reconstructed level is assigned to each pixel to obtain the final output R using the quantization encoding map, since E can reflect the original quantization level of each pixel. R is obtained from the following formula:
Finally, was reshaped as .
The second part performs semantic information feature extraction after underlying feature enhancement, exploiting texture-related information from multiple scales in a feature map containing rich texture details. Different from color features, texture features are not pixel-based features. It needs to be statistically calculated in an area containing multiple pixels, which is related to the statistical information of the spatial relationship. A gray-level co-occurrence matrix (GLCM) is used to capture the texture. The co-occurrence matrix is first generated by GLCM , and the texture information is represented by the contrast uniformity of the statistical description. In the processing of the feature map, it is first input into the two-dimensional quantization and counting operator to obtain the co-occurrence statistical features , and then use the multi-layer perceptron (MLP) and step-by-step averaging to generate the texture feature T of the processing area:
This paper also adopts the image pyramid structure, which is a structure that expresses images at multiple scales and interprets images at multiple resolutions. Enhanced texture features can be described from multiple scales. The designed pyramid structure adopts four parallel but different scale branch input feature maps. The feature map of each different branch is divided into a different number of sub-regions, and each sub-region passes through the texture feature extraction unit to utilize the corresponding texture representation of the region. Then, the obtained texture feature map of each branch is upsampled to the original size by nearest interpolation as the input map, and finally the outputs of the four branches are concatenated.
C.MULTIMODAL FEATURE REASONING MODULE
After obtaining the feature information of different modes, this feature information needs to be further processed to capture the complementary information between them. Therefore, we propose a feature fusion module called Multimodal Feature Reasoning Module (MFRM) to perform feature reasoning and information dissemination, as shown in Fig. 3. Finally, the information obtained by reasoning is upsampled to obtain segmentation results.
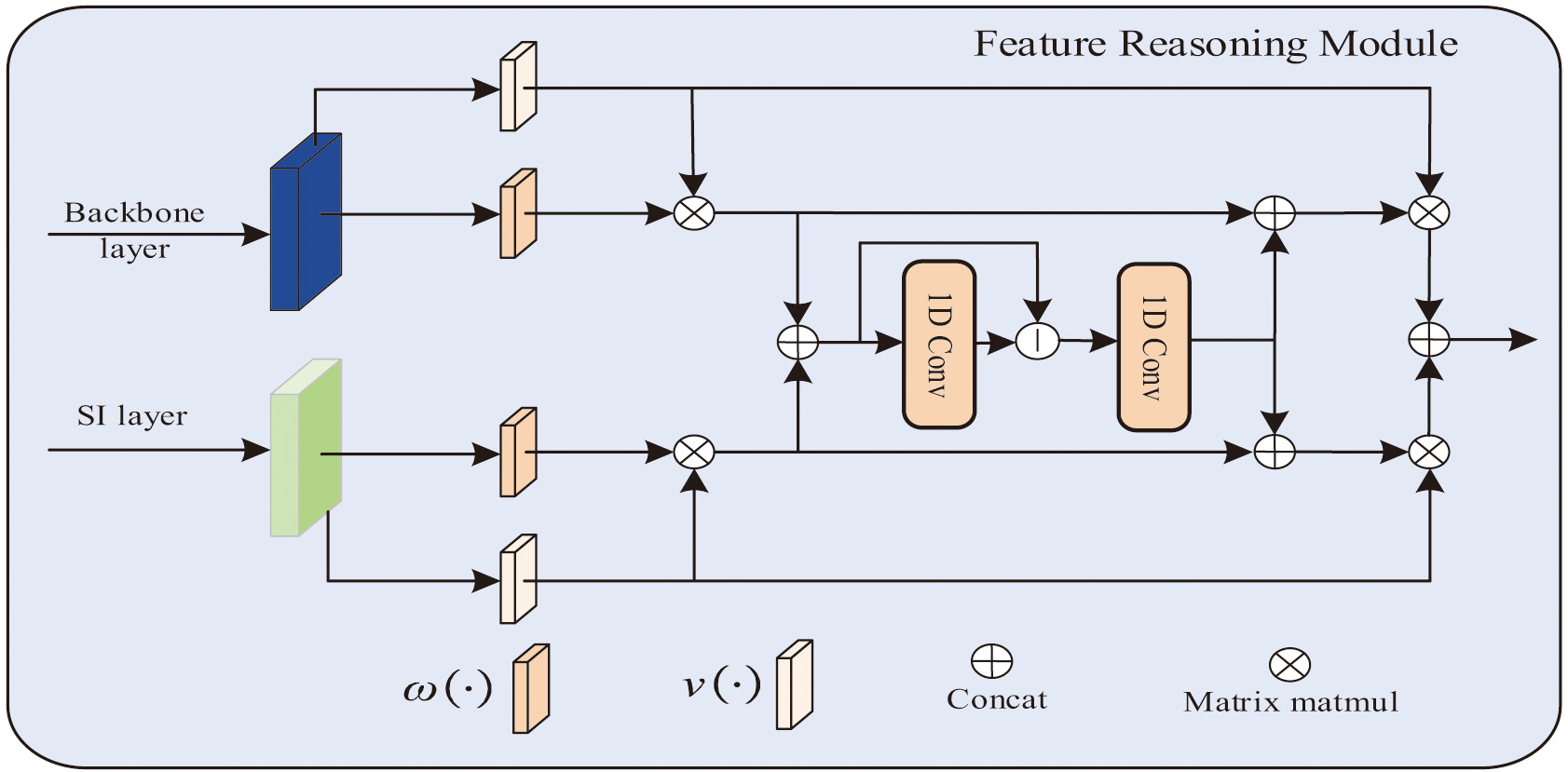 Fig. 3. Multimodal feature reasoning module.
Fig. 3. Multimodal feature reasoning module.
The features obtained by the given feature extraction backbone network and SIPM network are, respectively, represented as: , . In this paper, the input feature map is mapped from the spatial domain to the graph domain , where represents the number of nodes in the graph and represents the features contained in a node. In this way, pixels with similar features can be aggregated into a node as an anchor to generate a semantically aware graph feature. Specifically, the input features are mapped to image-domain features through two convolutional layers:
These represent the convolution operations used for graph projection and feature dimensionality reduction, and represent the learnable convolution kernels of each layer.
After projection, in order to learn the relationship between the semantic graph and the associated node features of the edge graph, we adopt graph convolution [44] to learn the edge weights corresponding to the features of each node to reason on the fully connected graph. The input is and the output is:
Among them, represents the identity matrix; represents the adjacency matrix; represents the update parameters.
The fused map to the original spatial domain via the projection matrix obtained in the mapping step to obtain new features . The obtained new features are passed through the upsampling network to obtain segmentation results.
IV.RESULTS
In this section, we will introduce the multimodal information fusion segmentation network structure proposed in this paper in detail. First, we will describe the overall structure of the proposed network in Section IV.A; then, in Sections IV.B and IV.C, we will discuss the innovations of this paper, SIPM and MFRM.
A.DATASET
In the experiments, the training and testing data sets are all from BraTS2018 and BraTS2019. As important public datasets for multimodal brain tumor segmentation, BraTS are used in MICCAI’s medical image segmentation competition. The data set will be added, deleted, or replaced in the competition every year to enrich the scale of the data sets. BraTS2018 and 2019 have 285 and 335 annotated brain tumor samples, respectively, they and are divided into highgrade glioma and low-grade glioma cases. Each case has 3D data of 4 different modes (Flair, t1, t1ce, t2). The datasets can differentiate three different brain tumor regions, namely Whole Tumor (WT), Tumor Core (TC), and Enhance Tumor (ET). The preprocessed datasets are composed of slices of different cases. During training, slices with tumors are used as data input, and all slices are cropped to reduce the amount of data and speed up training. The tested datasets are the brain tumor cases newly added in BraTS 2019 compared to BraTS 2018, a total of 50 labeled brain tumor samples.
B.EXPERIMENTAL DETAILS
In the preprocessing stage, the size of the source dataset is 240*240*155. In this paper, the data sets of all modalities are sliced. The size of each slice is the original 240*240, and the size of each slice after cropping is 160*160. And the original data will be standardized and normalized to realize the feature extraction and segmentation tasks of images in different modalities.
All the programs in this article are implemented under the PyTorch framework. The training process uses a GeForce RTX 3090 GPU. The optimizer during the experiment is Adam, the optimizer momentum is 0.9, and other parameters are default parameters. The initial learning rate, weight decay, and batch size are 0.001, 1e-4, and 32, respectively.
C.COMPARATIVE EXPERIMENT
First, model training and prediction are carried out on the method proposed in this paper. During the training process, the training loss (loss), IoU index of the training set and the test set, and the loss (val_loss) and val_IoU index of the test set are obtained, as follows. As shown in Fig. 4, for the previous training, the loss drops very quickly while the curve of the IoU index rises rapidly, and the subsequent training loss gradually decreases and becomes stable. Meanwhile, the rising curve of the IoU index gradually slows down and stabilizes, indicating that the training effect of the proposed method is ideal. To verify the superiority of the proposed multimodal information fusion segmentation network overall framework, this paper compares it with some classic brain tumor segmentation methods on the BraTS18,19 dataset. These methods include some classic segmentation methods FCN8 s, U-net, U-net++, Deep ResUnet (DRU) [45–48], and the experimental index comparison results are shown in Table I. The article uses several common indicators in image segmentation, Dice Score, Positive Predictive Value (PPV), and Hausdorff Score (HD). According to the experimental results, the method proposed in this paper has achieved considerable results in brain tumor segmentation.
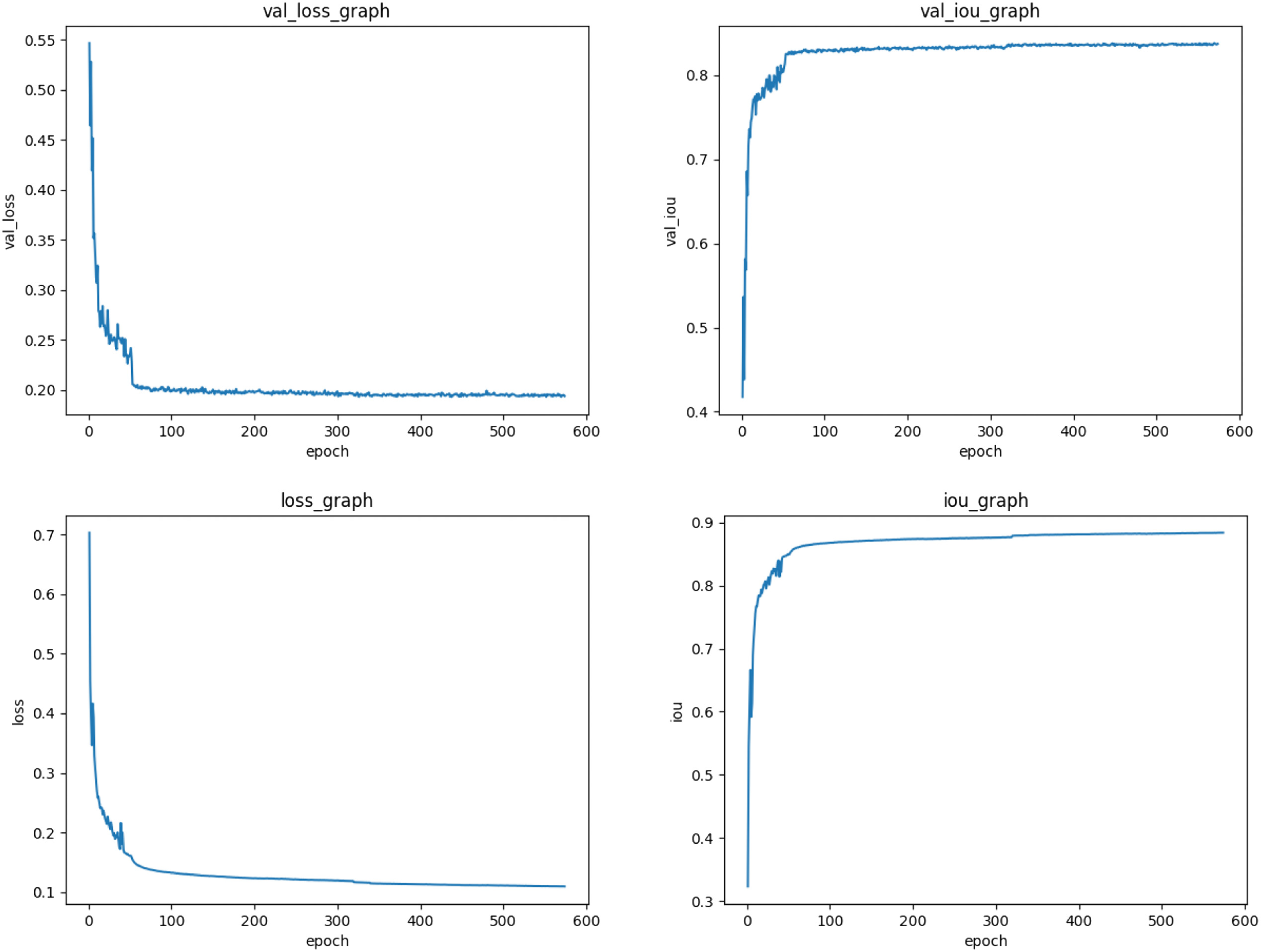 Fig. 4. Convergence of loss, val_loss, iou, val_iou during training.
Fig. 4. Convergence of loss, val_loss, iou, val_iou during training.
Table I. Comparison of segmentation indicators on Bra TS2018 and 2019
| Bra TS | WT | TC | ET | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Dice | HD | PPV | Dice | HD | PPV | Dice | HD | PPV | |
| FCN8s | 0.837 | 2.744 | 0.884 | 0.834 | 1.716 | 0.850 | 0.749 | 2.928 | 0.761 |
| U-net | 0.838 | 2.662 | 0.856 | 0.821 | 1.751 | 0.853 | 0.769 | 2.847 | 0.774 |
| Unet ++ | 0.842 | 2.638 | 0.866 | 0.827 | 1.722 | 0.850 | 0.774 | 2.815 | 0.792 |
| DRU | 0.847 | 2.589 | 0.888 | 0.834 | 1.693 | 0.860 | 2.779 | ||
| Ours | 0.781 | 2.778 | 0.812 | ||||||
It can be observed from the experimental results that the method proposed in this paper achieves considerable performance compared with the existing classical and advanced methods. Specifically, in terms of the Dice similarity coefficient score, the method based on the BraTS2018 and 2019 datasets achieved 87.2%, 87.6%, and 78.1% on the three segmentation tasks of whole tumor (WT), tumor core (TC) and enhancing tumor (ET), which are better than other classical segmentation methods, which are better than 2.50%∼3.50%, 1.3%∼5.5% and −0.1%∼3.2%, respectively. Compared with Unet, which also uses Resnet50 as the feature extraction network for semantic segmentation, this paper achieves a more effective improvement, among which the improvement in the tumor core is the most obvious, and its Dice coefficient is 5.5% better than that. At the same time, compared with other brain tumor segmentation networks using different modal fusions, the method in this paper has achieved improvements in the tumor core and enhanced tumors. The extraction of different features using different modalities in this paper has also been proven to be effective. The segmentation indicators of Dice, HD and PP have been improved. In addition, for the Dice index, as shown in the histogram of the segmentation index in Fig. 5, an intuitive comparison result is obtained, and the proposed method is superior to the classic advanced segmentation method.
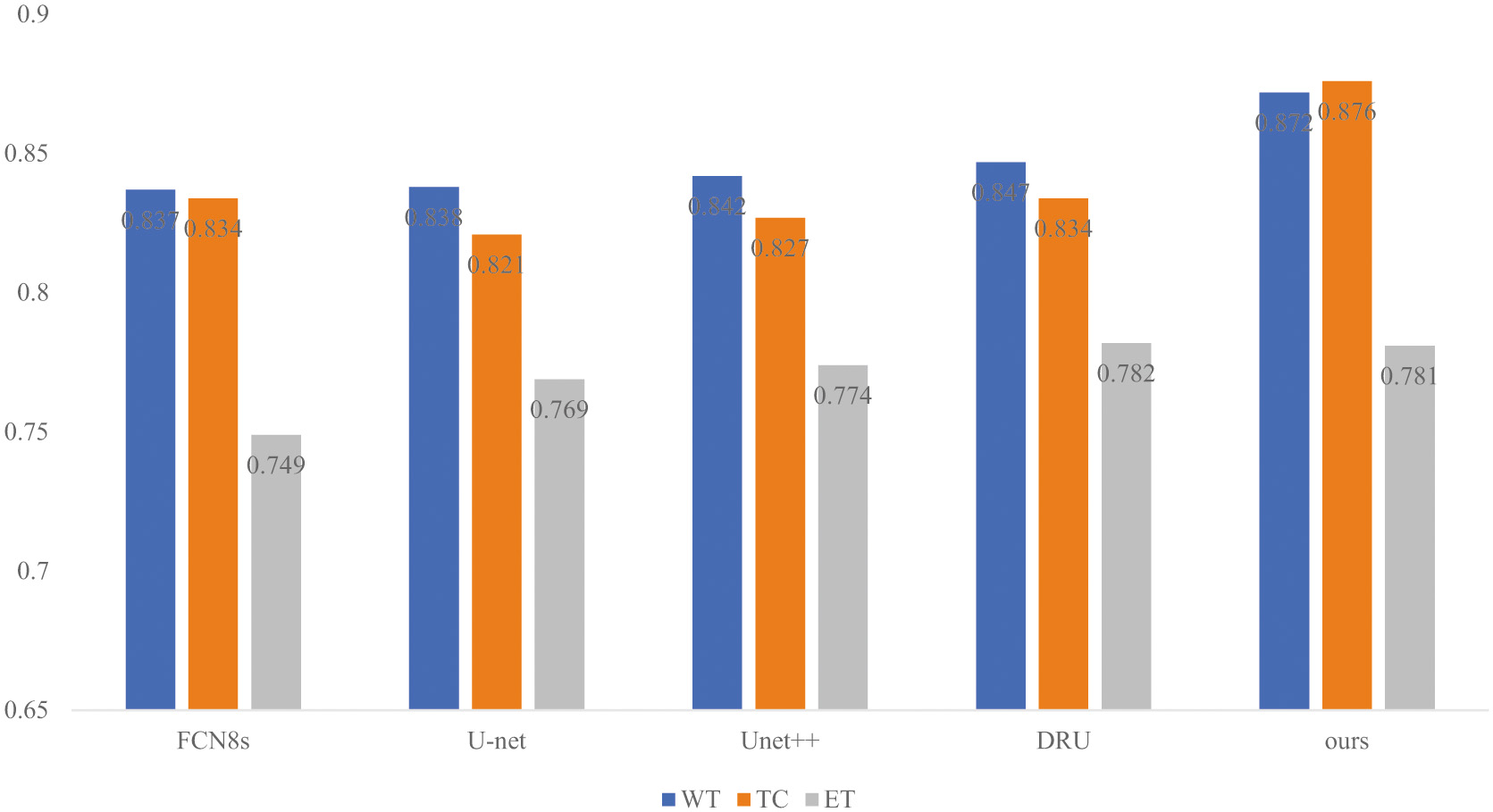 Fig. 5. Histogram of comparison experiment based on Dice indicator.
Fig. 5. Histogram of comparison experiment based on Dice indicator.
D.ABLATION EXPERIMENT
To further verify the importance and actual contribution of the backbone network used in this paper and the designed modules, relevant ablation experiments are carried out in this paper. The index comparison of the ablation experiment is shown in Table II, and the experimental results are shown in Fig. 7.
Table II. Comparison of ablation experiments of segmentation indicators on BraTS2018 and 2019
| Bra TS | WT | TC | ET | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Dice | HD | PPV | Dice | HD | PPV | Dice | HD | PPV | |
| Base | 0.825 | 2.780 | 0.839 | 0.820 | 1.753 | 0.846 | 0.748 | 2.938 | 0.754 |
| woSI-EE | 0.761 | 3.043 | 0.831 | 0.687 | 2.297 | 0.778 | 0.651 | 3.319 | 0.706 |
| woFRM | 0.807 | 2.944 | 0.808 | 0.782 | 1.939 | 0.816 | 0.712 | 3.168 | 0.700 |
| Ours | 0.872 | 2.584 | 0.905 | 0.876 | 1.511 | 0.956 | 0.781 | 2.778 | 0.812 |
In this paper, Resnet50 is used as the feature extraction base network (Base) for segmentation, and then the modules designed in this paper are added one by one on this basis for experiments, and SI-EE is added to the feature extraction network to improve and realize the enhancement of semantic information. And extraction, add an FRM module for feature fusion and feature reasoning. As shown in the performance index results in Table II, woSI-EE means that the SI-EE module is not added, and woFRM means that the FRM module is not added. According to the experimental results and data, it can be seen that the modules proposed in this paper can improve the Base Segmentation performance, among which the improvement of the FRM feature reasoning module on the above basis is the most significant. In addition, as shown in Fig. 6, based on the histogram comparison of the segmentation index Dice, the effectiveness of the proposed method for brain tumor segmentation can be obtained.
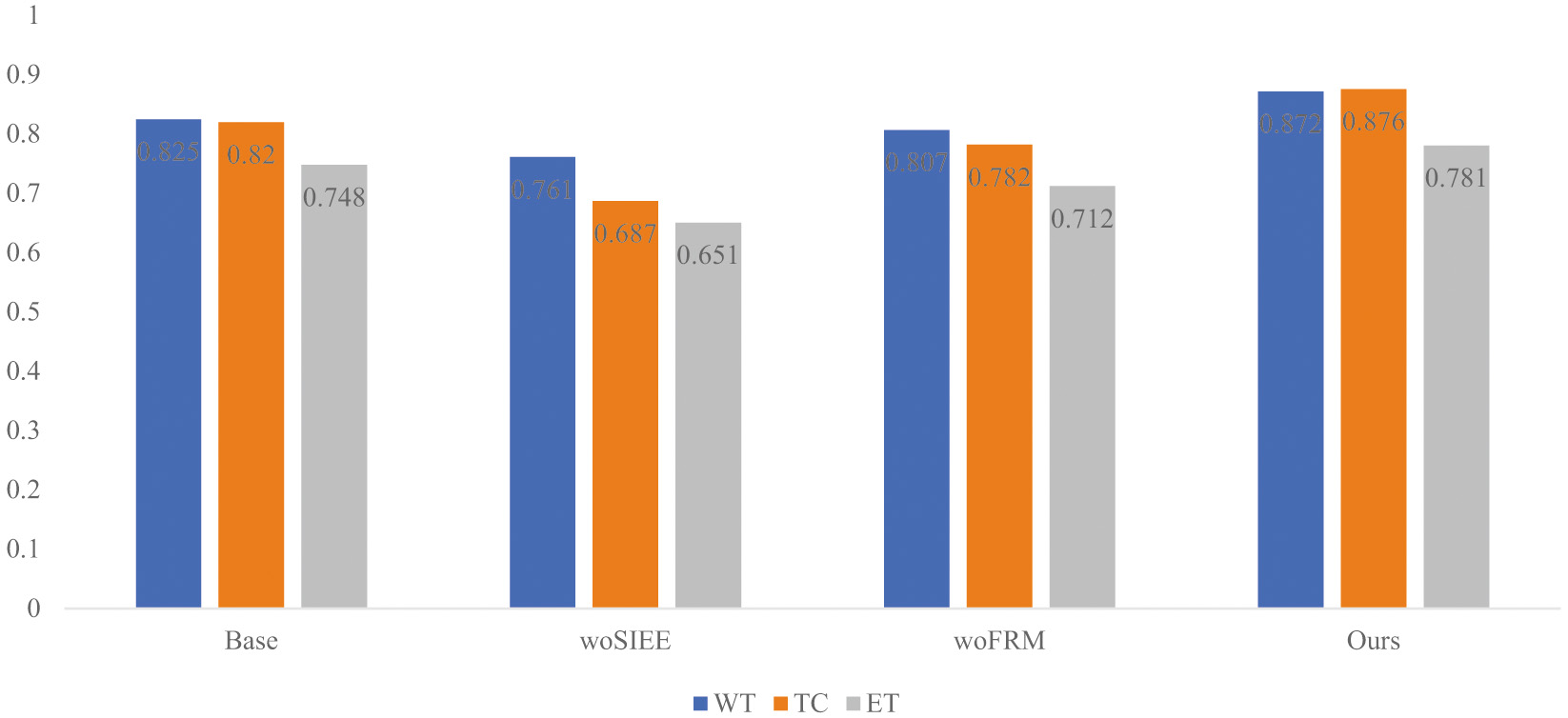 Fig. 6. Histogram of ablation experiment based on Dice index.
Fig. 6. Histogram of ablation experiment based on Dice index.
Figure 7 shows the visual images of the brain tumor segmentation obtained by each method in the ablation experiment. It can be seen from the figure that when the FRM module is not added, the edge segmentation of the brain tumor and the segmentation effect of the red area are not very good. At this time, under the action of the SI-EE module, compared with the Base, the segmentation results are greatly improved; When the SI-EE module is not added, the segmentation effect of the red area of the brain tumor is not very good, but it is easy to find that under the FRM module, the segmentation result of the edge area is closer to Ground Truth; in short, in the method proposed in this paper, the obtained brain tumor segmentation effect is smoother.
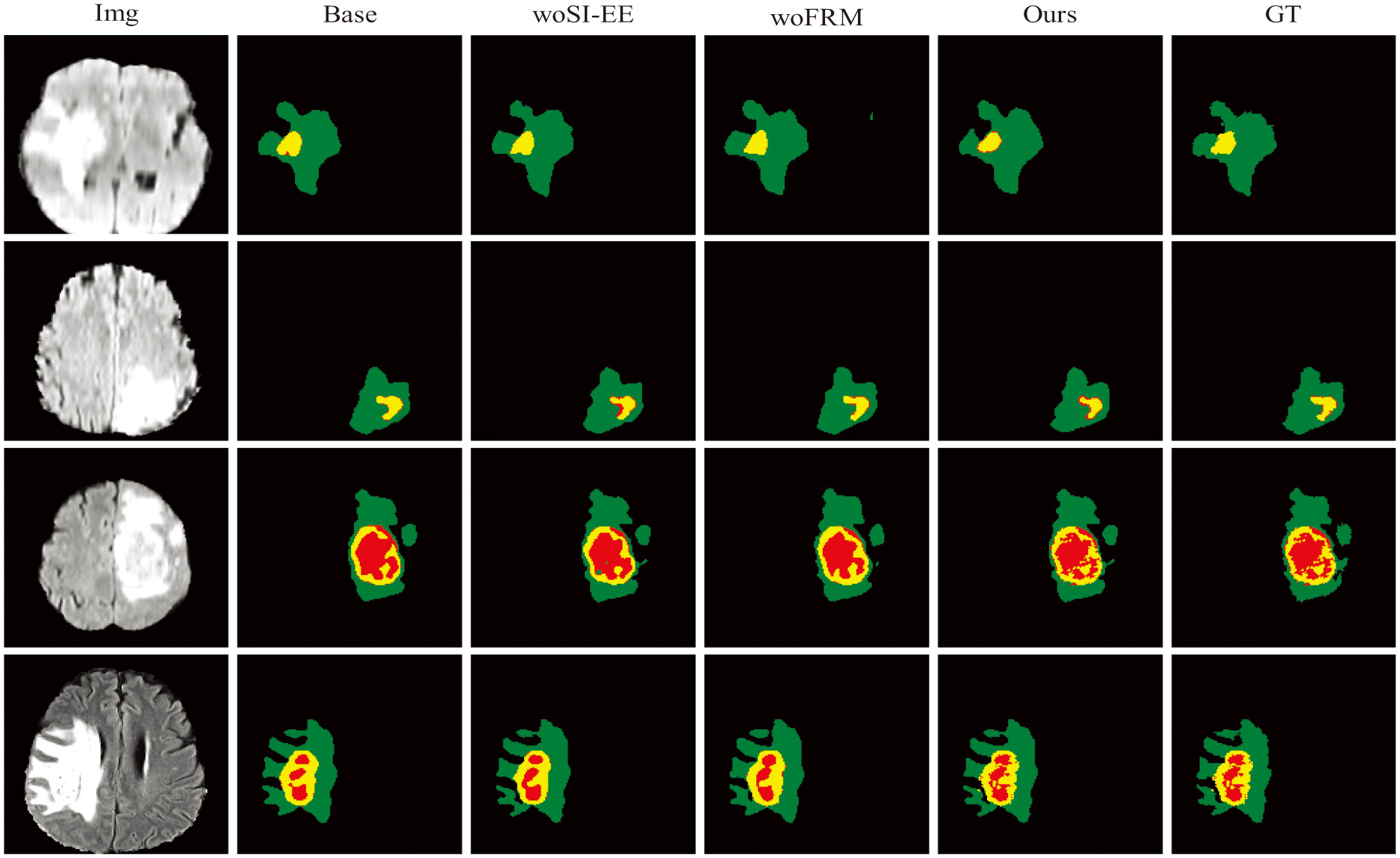 Fig. 7. Comparison of ablation experiments. Here green represents edematous areas, yellow represents enhancing tumors, and red represents necrotic and non-enhancing tumors.
Fig. 7. Comparison of ablation experiments. Here green represents edematous areas, yellow represents enhancing tumors, and red represents necrotic and non-enhancing tumors.
V.CONCLUSIONS
In this paper, we proposed a novel method of multimodal information fusion method for brain tumor segmentation. More comprehensive multimodal information from MRI was utilized to ensure accurate segmentation. Specifically, a Semantic Information Processing Module was introduced to achieve free Multiscale feature enhancement and extraction. The core of module formed a new SI-EE (Semantic Information Enhancement and Extraction) module. A new Multimodal Feature Reasoning Module was constructed to process both the backbone network feature information layer and semantic feature information layer. According to the experiments based on BraTS2018 and 2019 datasets, the method proposed in this paper achieved better results than the classic advanced segmentation methods.