I.INTRODUCTION
In the rapidly developing field of medical research and practice, the enormous amount of available information makes it challenging for healthcare personnel to keep up to date on the latest advancements. Medical text summarization emerges as a significant technique in this context, seeking to reduce complicated and vast medical literature into concise, useful summaries [1,2]. Medical text summaries, which use advanced natural language processing (NLP) techniques, not only help in efficient information acquisition but also play a significant part in clinical decision-making, medical education, and research endeavors. This unique technique has the potential to ease information exchange, resulting in improved patient care and scientific progress [3–5].
The use of medical text summarization marks a paradigm shift in information management in the healthcare sector. With the fast increase of biological literature and the continuous input of new research discoveries, healthcare practitioners have the arduous task of sifting through huge amounts of data to extract relevant and practical insights [6,7]. Machine learning and NLP techniques are used to extract core concepts, identify key evidence, and condense complicated medical texts into succinct summaries. This not only speeds knowledge acquisition but also allows practitioners to make more informed judgments by focusing on the most relevant information. The role of medical text summarization is becoming increasingly significant in developing a more efficient and knowledge-driven healthcare system [8].
Currently of modern technology, the use of transformers in medical text summarization is a game changer [9,10]. Transformers, as demonstrated by models like as BART (Bidirectional Autoregressive Transformer), BERT (Bidirectional Encoder Representations from Transformers), GPT (generative pretrained transformer), and T5, have transformed NLP by capturing rich contextual links within textual input. In the medical field, these transformer-based models excel at understanding the intricacies of sophisticated scientific jargon, allowing them to distill lengthy medical texts with surprising precision. Using attention processes and contextual embeddings, these models can distinguish significant material, highlight essential discoveries, and provide coherent summaries that preserve the core of the original content. This unique approach not only speeds up and optimizes information extraction, but it also has great promise for improving clinical decision support systems, medical research, and healthcare professional training materials [11,12]. Transformers continue to progress, and their application to medical text summarization ushers in a new age of accuracy and efficiency in dealing with the ever-expanding medical information environment.
Fine-tuned transformers provide a customized approach to medical text summarization, using the capabilities of pretrained models while tailoring them to the unique requirements of the healthcare domain. These models, which were trained on large-scale medical datasets, show a better grasp of medical terminology, context, and domain-specific nuances. The fine-tuning approach allows for customization, allowing the transformer to improve its summarizing abilities depending on the specific characteristics of medical literature. Fine-tuned transformers excel in producing brief and accurate summaries that accurately handle the complexities of healthcare discourse by emphasizing medical terminology, expert vocabulary, and contextually relevant information. This accuracy not only allows for more effective information extraction by healthcare professionals, but it also has the potential to profoundly affect crucial areas such as clinical decision-making, research synthesis, and medical education. The use of fine-tuned transformers in medical text summarization demonstrates a subtle and sophisticated approach to handling the quantity of information in the ever-changing world of medical research and practice. Fine-tuned transformers bring a tailored precision to medical text summarization by adapting pretrained models to the specific nuances of the healthcare domain. This customization ensures a heightened understanding of medical terminology, enabling the generation of more accurate and contextually relevant summaries.
The key contribution of the research include
- a)Exploring the extractive text summarization aspects of medical and drug discovery.
- b)Collection of abstracts collected from PubMed for various domains of medical and drug discovery such as drug, Naïve, Covid, influenza, Clinical text, Covaxin, vaccine, microbiotia, and computational.
- c)Using BART, T5, LexRank, and TexRank for the analysis of the dataset to perform extractive text summarization.
Our study introduces several key advancements that go beyond standard approaches. First, we have implemented a fine-tuning process on transformer models specifically for medical texts, ensuring that these models are adept at handling the specialized language and terminology used in drug and vaccine literature. This fine-tuning process allows the model to capture the nuanced relationships and contextual information crucial for accurate summarization in this domain. Second, we incorporated a customized attention mechanism that prioritizes medical terms and their related concepts, improving the relevance and precision of the summaries generated. Additionally, our approach includes a unique algorithm designed to filter and prioritize sentences that contain critical drug and vaccine information, which is particularly important for ensuring that the summaries are both concise and informative. These innovations demonstrate our focus on tailoring existing technologies to meet the specific demands of the drug and vaccine fields, providing a more effective tool for medical professionals and researchers.
The rest of the of the article consists of the related and prior works in the domain under Section II, followed by Section III mentioning the details of the proposed model for handling the drug and medical data for extractive summarization. Later sections provide the details of experimentation, results, and analysis. The article is concluded with a summary and future directions.
II.RELATED WORK
Text summarization can be categorized into two main types: extractive and abstractive. In extractive summarization, the summary is created by selecting and combining sentences or segments from the original document. Each sentence is typically ranked based on its importance, and then these selected sentences are rearranged to form the summary while adhering to grammatical rules. On the other hand, abstractive summarization involves interpreting the original document and generating concise, meaningful sentences that capture its essence. This approach requires a deeper semantic understanding of the document to craft a summary that is easily comprehensible to humans. The works reported in this section discusses the various extractive text summarization models.
Moradi et al. [13] discuss the advantages of contextualized embeddings related to biomedical text summarization. Unlike context-free embeddings, contextualized embeddings consider the context in which words appear, enabling a more nuanced representation of semantic and syntactic information. This approach addresses challenges related to incorporating domain knowledge, as the embeddings are learned from large corpora in an unsupervised manner. This approach allows for the effective quantification of shared context without the need for annotating or maintaining biomedical knowledge bases. The method focuses on capturing context, quantifying relatedness, and selecting informative sentences, offering a potential advancement in the field of biomedical NLP.
He et al. [14] introduce an approach to address challenges that are specific to multi-label classification of clinical texts. Two main challenges are identified: label class imbalance and the need for extracting semantic features. To tackle the first challenge, the authors propose a method to enrich label representations with supplementary information and leveraging co-occurrence relationships between labels. The second challenge is also addressed to an extent by the authors [14]. Clinical text representation learning involves fine-tuning Bio-BERT for specific medical domain tasks, extracting fine-grained semantic information. Prediction scores for each label are generated through mapping and processing layers, determining the most relevant labels for a given text.
The extractive summarization framework proposed by Li et al. [15] aims to enhance the summarization of academic articles in natural sciences and medicine by incorporating comprehensive section information. The framework involves three key steps: designing section-related questions, labeling ground truth answer spans, and training the model based on large-scale language models.
The study in [15a] proposed a supplementary extractive summarization framework that demonstrated the effectiveness of contrastive learning in an effort to improve model performance. This was accomplished by placing an emphasis on the difference between information that is important and information that is not relevant.
Ozyegen et al. [16] discuss an online chat service connecting patients with board-certified medical doctors, offering a novel approach to healthcare by allowing remote communication of symptoms and health concerns. Like telehealth, this service aims to provide quality healthcare, eliminating barriers like time off work and high medical costs. Particularly beneficial during the COVID-19 pandemic, it enhances medical access without in-person visits, potentially saving resources and reducing infections. In the context of this online medical chat service, the paper’s primary goal is to develop a mechanism that highlights crucial words and short segments in patient messages, streamlining doctors’ responses by focusing on essential information. The objective is to expedite the reading and response time for doctors. Zhu et al. [17] present a comprehensive framework for entity recognition, relation extraction, and attribute extraction for the corpus obtained from HwaMei Hospital in Ningbo, China, that involves 1200 manually annotated electronic medical records. The study [17] employs neural NLP models like BERT.
Szeker et al. [18] introduce a versatile text mining approach for extracting numerical test results and their descriptions from free-text echocardiography reports, departing from regex-based methods. It employs corpus-independent techniques, automatically identifying expressions in medical texts and associating them with numerical measurements. Fuzzy matching is used to identify candidate terms, enabling flexible handling of typos and abbreviations. The method [18], tested on over 20,000 echocardiography reports, proves efficient for rapidly processing large datasets, supporting medical research, and swiftly verifying patient selection criteria for clinical trials.
Du et al. [19] present a domain-aware language model for summarization in medical field and introduced a position embedding for sentences that captures structural information. Xie et al. [20] introduce a method for biomedical extractive summarization by integrating medical knowledge into pretrained language models (PLMs). This approach employs a lightweight training framework, the knowledge adapter, which utilizes distant annotations for efficient integration of medical knowledge. Through generative and discriminative training, the method masks various elements in input sentences, reconstructing missing tokens, and predicting element labels. Trained adapters, capturing domain knowledge, are then fused into PLMs, enhancing their ability to focus on domain-specific tokens during fine-tuning for improved extractive summarization [20]. The works [21,22] also try to address the biomedical summarization aspects using frequent item sets mining, using the K-means algorithm [21] and a shuffled leaping algorithm [22].
Han et al. [23] introduce a framework for extractive summarization addressing issues of redundant content and long-range dependencies. The proposed topic model transforms word-level topics into sentence-level information, utilizing a heterogeneous graph neural network for sentence selection. A dynamic memory unit is incorporated to prevent repetitive features from influencing the model’s decision. The framework employs reinforcement learning for parameter updates, mitigating training-testing discrepancies. The authors [24] introduces a novel approach by integrating BERT with an Attention-based Encoder-Decoder for enhanced summarization of lengthy articles. The proposed Chunk-based Framework partitions documents into sentence-level chunks, utilizing a fusion of BERT and other transformers. An algorithm seamlessly incorporates BERT into the summarization process, demonstrating robust performance compared to state-of-the-art methods.
Bano et al. [25] introduce a medical specialty prediction framework using pretrained BERT from medical question text. However, limitations include data quality dependence and variations in predictive difficulty among medical specialties, urging caution in practical applications. Future work aims to address these challenges and enhance overall predictive performance across medical specialties. Kim et al. [26] introduce a multi-document biomedical text summarizer employing concept mapping and item set mining for topic discovery.
Earlier studies primarily employed traditional summarization methods, such as TF-IDF and LDA, which often struggled to capture the complex contextual nuances required for effective medical text summarization. Our research improves upon this by utilizing advanced fine-tuned transformer models, specifically BART and T5, which are adept at handling the intricate language of medical texts. These models significantly enhance the contextual understanding through their self-attention mechanisms, leading to more coherent and relevant summaries. Unlike generic models used in past research, our approach involves fine-tuning transformers on medical-specific datasets, which results in more accurate and contextually appropriate summaries. Additionally, our models are better equipped to handle long and complex documents, maintaining coherence and fluency throughout. The quality of the summaries is further evaluated using both ROUGE and specialized metrics like cosine similarity, providing a thorough assessment. This approach also offers scalability and efficiency, making it highly suitable for processing large volumes of medical data in research settings.
III.PROPOSED WORK
Transformers such as BERT have significantly improved the performance of various NLP tasks by leveraging their ability to capture a wide range of linguistic features. These models are adept at handling diverse downstream tasks thanks to their robust representations of language. One common approach to adapting these models to specific tasks is task adaptation, where the model is fine-tuned using examples from the target task to achieve optimal performance. However, this approach treats each task’s model as separate entities, which complicates the sharing of knowledge across tasks.
In contrast, multi-task learning (MTL) offers a more integrated solution by combining multiple related tasks into a single neural model. This allows for seamless knowledge-sharing between tasks within the same model architecture. While traditional transfer learning methods can incorporate BERT embeddings into MTL setups, they may not efficiently leverage the fine-tuned embeddings tailored to specific tasks. Therefore, leveraging fine-tuned embeddings rather than generic embeddings through MTL can lead to more effective knowledge-sharing and improved performance across tasks.
A.DATA COLLECTION
This article initially prepares the dataset required to carry out the tasks required to perform summarization. A total of 1370 articles are collected from different domains from PubMed. The articles belong to the domains such as Drug, Naïve, Covid, Influenza, Clinical text, Covaxin, Vaccine, Microbiotia, and Computational. The classification of the data collected is shown in Table I. The selected topics are highly relevant to current medical research and public health. These areas generate significant volumes of literature and summarizing them ensures that critical information is quickly accessible to researchers and healthcare professionals. This selection supports the efficient dissemination of knowledge, fostering advancements in treatment, vaccine development, and public health strategies.
Table I. Classification of articles collected for the summarization task
| Domain | No of abstracts collected |
|---|---|
| Drug | 196 |
| Naïve | 197 |
| Covid | 178 |
| Influenza | 70 |
| Clinical text | 198 |
| Covaxin | 192 |
| Vaccine | 141 |
| Microbiotia | 148 |
| Computational | 50 |
Text summarization is a vital task in NLP, and the proposed model aims to enhance extractive summarization using transformer-based architectures. Leveraging the strengths of transformers in capturing contextual information, the model seeks to address redundancy and coherence issues in existing methods.
In our exploration of deciphering human emotions embedded in text, we’ve opted for BERT, a pretrained transformer model, as the foundation for our research in extractive text summarization, utilizing fine-tuned transformers. BERT’s superiority over models like Long Short Term Memory (LSTM) and Convolutional Neural Networks (CNNs) stems from its finesse in fine-tuning, leveraging pretrained parameters adaptable to various domains. One of BERT’s standout features is its multi-head attention mechanism, which allows for simultaneous focus on different aspects of the input text. This capability enables nuanced emotion recognition by grasping relationships and long-range dependencies within the text. Moreover, BERT incorporates layer normalization for stable training and excels in utilizing pretrained word embeddings, capturing subtle semantic nuances crucial for accurate emotion recognition. This comprehensive approach establishes BERT as the preferred choice, delivering heightened accuracy and deeper insights into the realm of human sentiment within textual data.
B.SELF-ATTENTION MECHANISM
The self-attention mechanism, which plays a crucial role in models like BERT, is valued for its capacity to grasp contextual nuances and manage long-range dependencies within input sequences. Unlike traditional sequential models, self-attention permits simultaneous consideration of all positions within a sequence, facilitating parallelization during training and adaptability to sequences of varying lengths. This bidirectional mechanism not only addresses issues like vanishing or exploding gradients but also effectively captures contextual relationships in both forward and backward directions. In essence, self-attention significantly enhances the efficiency and performance of models in NLP tasks. BERT’s self-attention mechanism operates based on the scaled dot-product attention.
The proposed architecture for utilizing the BART model for extractive text summarization on drug and vaccine data involves several interconnected modules designed to process input sequences, capture contextual information, and generate concise summaries as shown in Fig. 1. At its core, BART leverages the transformer architecture, which consists of encoder and decoder layers, each comprising various submodules to handle different aspects of text summarization.
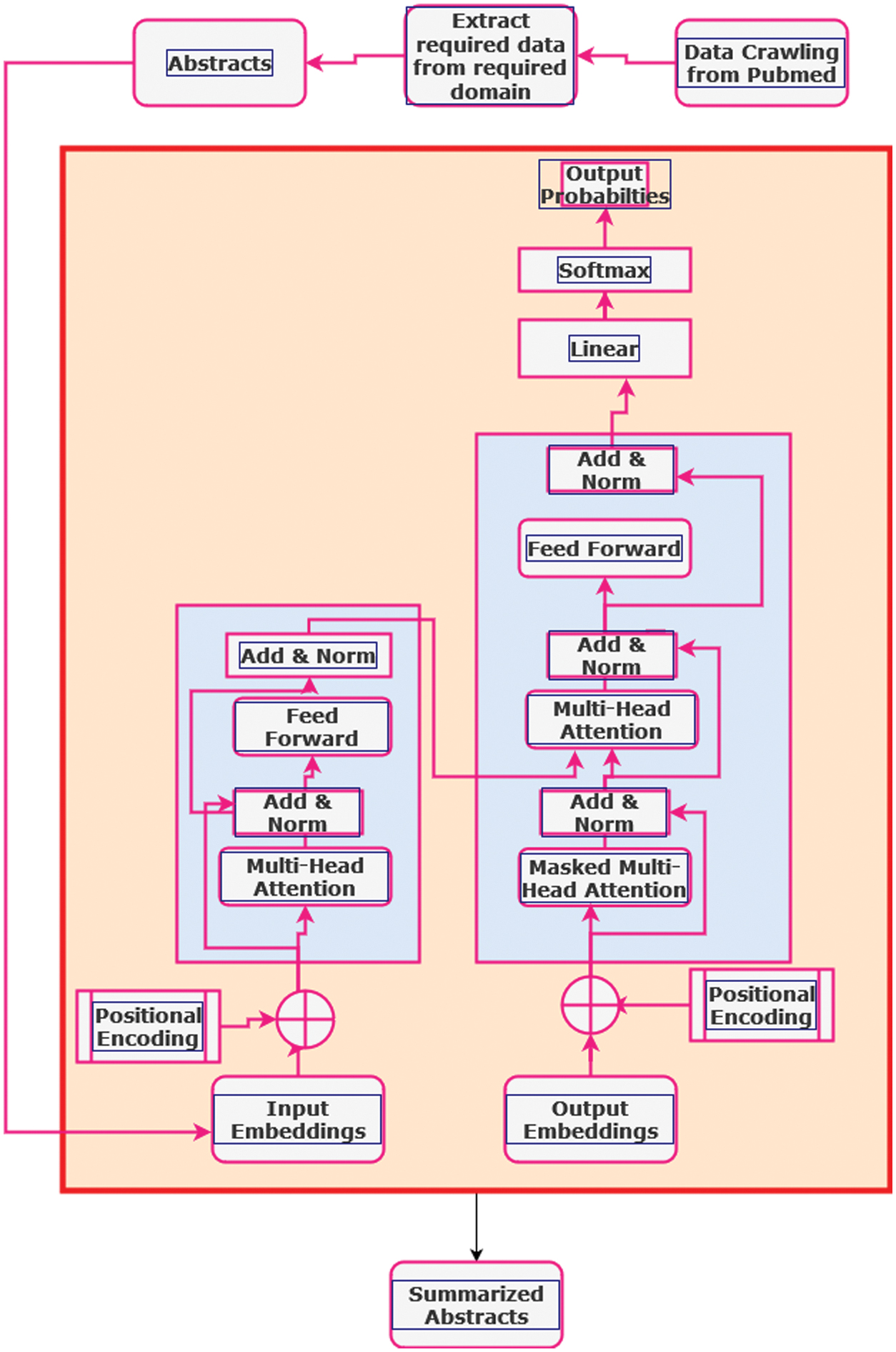 Fig. 1. Proposed model for the extractive text summarization.
Fig. 1. Proposed model for the extractive text summarization.
1.INPUT REPRESENTATION
The initial stage in the BART architecture is to encode the input text into token embeddings. An embedding layer maps each token in the input sequence to a multidimensional vector space. This embedding layer turns the discrete token indices into continuous-valued vectors, allowing the model to handle textual input. This may be expressed mathematically as follows:
where X represents the input token sequence and n is the length of the sequence.2.ENCODER LAYERS
The input token embeddings are subsequently delivered through many encoder layers. Each encoder layer has two main submodules: the multi-head self-attention mechanism and the position-wise feed forward neural network. The self-attention strategy allows the model to recognize links between the characters in the input sequence. It computes attention ratings for each pair of tokens and generates weighted representations accordingly. Attention scores are determined using the scaled dot-product attention method, which gives
where Q, K, and V are the query, key, and value matrices, respectively, and dk is the dimension of the key vectors.3.POSITION-WISE FEEDFORWARD NETWORK
After computing the self-attention ratings, the resultant representations are fed into a position-wise feedforward neural network (FFN). This FFN consists of two linear transformations separated by a Rectified Linear Unit (ReLU) activation function. The FFN aids in capturing complicated patterns and relationships within the input sequence, allowing the model to learn rich representations of textual material.
4.DECODER LAYERS
After the input sequence is encoded, the decoder layers use the encoded representations to construct the summary. Each decoder layer consists of two main submodules: masked multi-head self-attention and cross-attention. The masked multi-head self-attention mechanism attends to all positions in the input sequence, but only permits each position to attend to the positions before it. This prohibits the model from accessing future data during training, ensuring that the generation process stays autoregressive.
5.CROSS-ATTENTION MECHANISM
In addition to masked self-attention, the decoder layers provide a cross-attention mechanism. This method allows the model to focus on important information in the encoded input sequence while creating the summary. It enables the decoder to focus on certain sections of the input sequence based on the context supplied by the summary generating process.
6.OUTPUT GENERATION
Finally, the decoder layers’ output is transmitted via a linear layer, followed by a softmax function, which computes the probability distribution across tokens in the vocabulary. During extractive summarization, the model picks phrases or segments from the input sequence using the probabilities assigned to each token. This selection procedure produces a summary that highlights the most important information found in the supplied text.
The BART model can successfully handle drug and vaccine-related textual data by combining these interrelated modules and generating extractive summaries that capture the relevant information within the input sequences. The design takes use of the transformer’s capacity to capture long-term dependencies and contextual information, making it ideal for text summarizing jobs in specialized sectors such as medication and vaccine research.
IV.RESULT AND DISCUSSION
The comparative analysis of various summarization models, including BART, T5, TexRank, and LexRank, provides valuable insights into their performance across different domains. The results of the data collected are presented in this section.
BART demonstrates consistent and moderate effectiveness in capturing key linguistic features, particularly excelling in the domain of clinical text. Its ability to maintain strong similarity between summaries and abstracts highlights its capacity to generate coherent summaries. While T5 exhibits lower ROUGE scores compared to BART, it still demonstrates reasonable performance, albeit with less consistency across domains. Despite facing challenges in capturing certain linguistic elements, T5 shows potential for generating informative summaries. TexRank and LexRank models showcase moderate to stable performance across domains, effectively capturing the essence of the source documents in their summaries. However, it is important to note that all models display variations in performance across different domains, indicating the necessity for further investigation into domain-specific optimizations to enhance summarization quality.
Overall, this analysis provides valuable insights for researchers and practitioners in the field, offering a nuanced understanding of the strengths and weaknesses of each summarization model and highlighting potential avenues for future research and development. Figure 2 shows an example of summarization. Analyzing the summaries generated by each model – BART, LexRank, and T5 – provides insights into their respective approaches and effectiveness in capturing the key information from the abstract. Starting with the BART summary, it effectively highlights the main findings of the study, emphasizing the role of myosteatosis as a potential predictor of new-onset diabetes mellitus after kidney transplantation. The summary also provides essential details such as the prevalence of impaired glucose tolerance and new-onset diabetes mellitus among the patient cohort, along with the relevant measurements of psoas muscle index and average total psoas density. However, the BART summary lacks some context, such as the time frame of the study and the significance of the findings, which could be important for readers seeking a comprehensive understanding of the research.
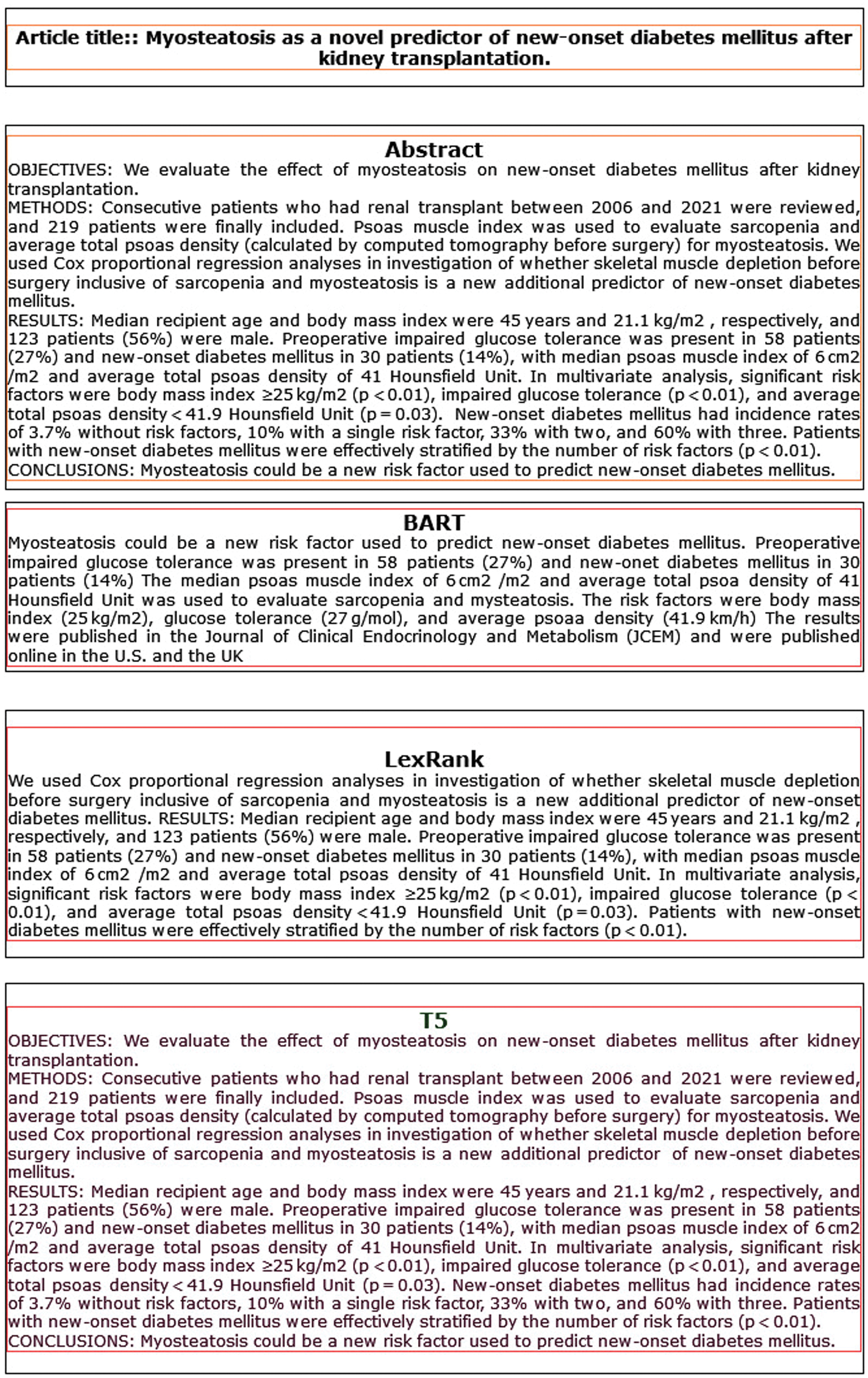 Fig. 2. Example text summarization.
Fig. 2. Example text summarization.
Moving on to the LexRank summary, it presents a similar overview of the study, focusing on the evaluation of skeletal muscle depletion, including sarcopenia and myosteatosis, as predictors of new-onset diabetes mellitus. Like the BART summary, it provides details regarding patient characteristics, prevalence rates of impaired glucose tolerance and new-onset diabetes mellitus, and significant risk factors identified through multivariate analysis.
However, it also lacks contextual information such as the time frame and publication details of the study, which could limit its usefulness for readers seeking additional context or background information. Finally, the T5 summary mirrors the content of the original abstract, presenting the study objectives, methods, results, and conclusions in a concise and coherent manner.
It effectively communicates the key findings of the study, including the evaluation of myosteatosis as a predictor of new-onset diabetes mellitus and the identification of significant risk factors through multivariate analysis. Additionally, it provides information on the prevalence rates of impaired glucose tolerance and new-onset diabetes mellitus among the patient cohort, along with relevant measurements of psoas muscle index and average total psoas density. Overall, the T5 summary offers a comprehensive overview of the study findings and is likely to be informative for readers seeking a concise summary of the research.
Each model – BART, LexRank, and T5 – provides a summary of the study on the relationship between myosteatosis and new-onset diabetes mellitus after kidney transplantation. While each summary captures the main findings and key details of the study, they vary in terms of completeness and contextual information. The T5 summary stands out for its comprehensive coverage and clear presentation of the study objectives, methods, results, and conclusions. However, all summaries could benefit from including additional context, such as the time frame and publication details of the study, to provide readers with a more thorough understanding of the research.
A.RESULTS USING BART
Table II and Fig. 3 present ROUGE-1, ROUGE-2, and ROUGE-L scores for the BART model across various domains. Overall, the model’s performance is fairly consistent across domains, with ROUGE-1 scores ranging from 0.19 to 0.21, ROUGE-2 scores ranging from 0.21 to 0.24, and ROUGE-L scores ranging from 0.69 to 0.74.
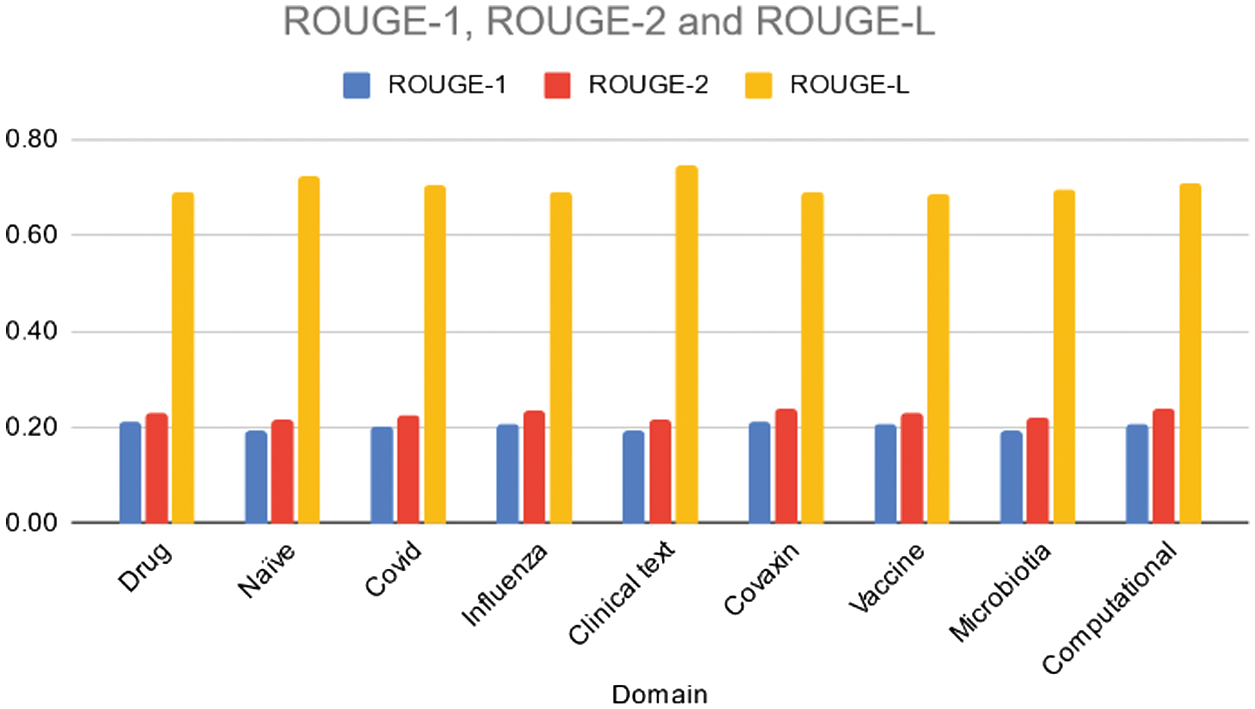 Fig. 3. Rouge 1/2/L scores of the various domains using BART model.
Fig. 3. Rouge 1/2/L scores of the various domains using BART model.
Table II. ROUGE scores of various domains using BART model
| Domain | ROUGE-1 | ROUGE-2 | ROUGE-L |
|---|---|---|---|
| 0.21 | 0.23 | 0.69 | |
| 0.19 | 0.21 | 0.72 | |
| 0.20 | 0.23 | 0.71 | |
| 0.21 | 0.23 | 0.69 | |
| 0.19 | 0.22 | 0.74 | |
| 0.21 | 0.24 | 0.69 | |
| 0.21 | 0.23 | 0.69 | |
| 0.19 | 0.22 | 0.69 | |
| 0.21 | 0.24 | 0.71 |
The highest ROUGE-L score is observed in the “Clinical text” domain, indicating better fluency and coherence compared to other domains. While there are slight variations in performance, the model generally demonstrates moderate effectiveness in capturing unigrams, bigrams, and longest common subsequences across different topic areas. Further analysis could explore specific reasons for variations and potential areas for improvement in domain-specific summarization tasks.
Table III and Fig. 4 display cosine similarity scores generated by the BART model for data summaries across various domains. Across all domains, the BART model demonstrates strong similarity between summaries and abstracts, with scores ranging from 0.85 to 0.87. This indicates that the summaries effectively capture the essence of the information presented in the abstracts of the documents. Similarly, the model exhibits consistent similarity between abstracts and titles, ranging from 0.71 to 0.73, suggesting that the abstracts accurately convey the core content outlined in the titles. Furthermore, there is moderate to high similarity between summaries and titles, with scores ranging from 0.75 to 0.77, indicating that the summaries generally reflect the main content outlined in the titles. Overall, the BART model demonstrates consistent performance across domains in aligning summaries with abstracts and titles, underscoring its capability to generate coherent and relevant summaries. Further analysis could explore potential domain-specific nuances and areas for improvement to enhance summarization quality in specific topics.
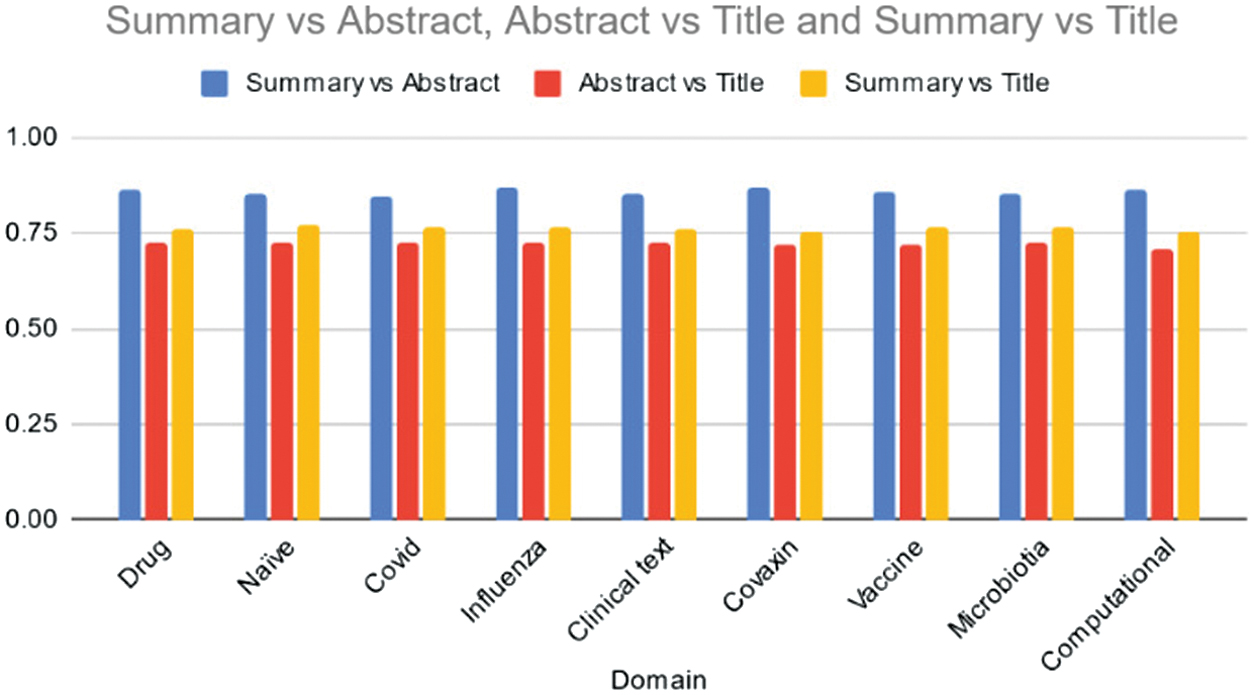 Fig. 4. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using BART model.
Fig. 4. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using BART model.
Table III. Average cosine similarity of the summary vs abstract, abstract vs title, and summary vs title across various domains using BART model
| Domain | Summary vs abstract | Abstract vs title | Summary vs title |
|---|---|---|---|
| Drug | 0.86 | 0.72 | 0.76 |
| Naïve | 0.85 | 0.72 | 0.77 |
| Covid | 0.85 | 0.72 | 0.77 |
| Influenza | 0.87 | 0.73 | 0.77 |
| Clinical text | 0.85 | 0.72 | 0.76 |
| Covaxin | 0.87 | 0.72 | 0.75 |
| Vaccine | 0.86 | 0.72 | 0.76 |
| Microbiotia | 0.85 | 0.73 | 0.77 |
| Computational | 0.86 | 0.71 | 0.75 |
B.RESULTS USING T5 MODEL
Table IV and Fig. 5 outline the ROUGE-1, ROUGE-2, and ROUGE-L scores achieved by the T5 model across different domains. Across the board, the T5 model exhibits lower scores compared to the BART model, indicating potential differences in summarization quality between the two models. Notably, ROUGE-1 scores range from 0.11 to 0.13, ROUGE-2 scores range from 0.10 to 0.13, and ROUGE-L scores range from 0.82 to 0.86. The “Influenza” and “Clinical text” domains have the lowest ROUGE-1 and ROUGE-2 scores, suggesting challenges in capturing unigrams and bigrams effectively in these topics. Overall, the T5 model’s performance appears to be less consistent across domains compared to the BART model, indicating potential areas for improvement or domain-specific tuning in summarization tasks.
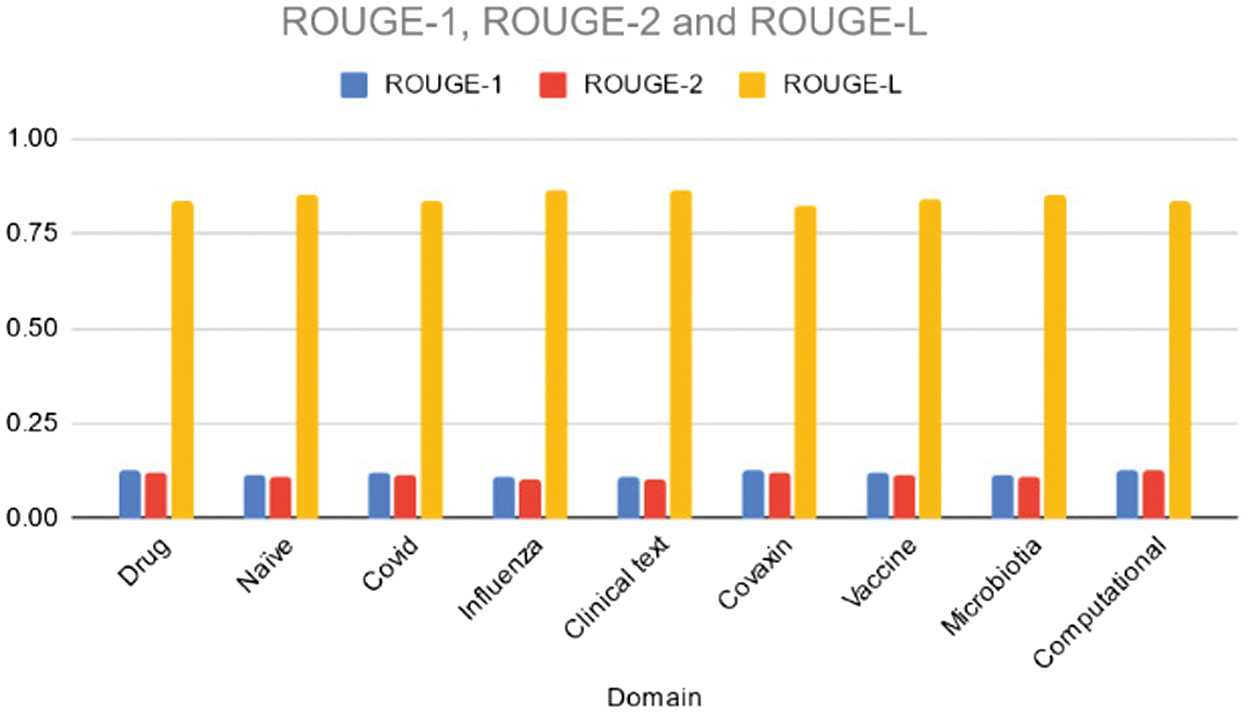 Fig. 5. Rouge 1/2/L scores of the various domains using T5 model.
Fig. 5. Rouge 1/2/L scores of the various domains using T5 model.
Table IV. ROUGE scores of various domains using T5 model
| Domain | ROUGE-1 | ROUGE-2 | ROUGE-L |
|---|---|---|---|
| Drug | 0.13 | 0.12 | 0.83 |
| Naïve | 0.12 | 0.11 | 0.85 |
| Covid | 0.12 | 0.12 | 0.84 |
| Influenza | 0.11 | 0.10 | 0.86 |
| Clinical text | 0.11 | 0.10 | 0.86 |
| Covaxin | 0.13 | 0.12 | 0.82 |
| Vaccine | 0.12 | 0.11 | 0.84 |
| Microbiotia | 0.11 | 0.11 | 0.85 |
| Computational | 0.13 | 0.13 | 0.83 |
Table V and Fig. 6 present cosine similarity scores derived from the T5 model for data summaries across different domains. Across all domains, the T5 model demonstrates relatively high similarity scores between summaries and abstracts, with values ranging from 0.82 to 0.83. This suggests that the generated summaries effectively capture the essence of the information presented in the abstracts of the documents. Moreover, the model shows consistent similarity between abstracts and titles, ranging from 0.71 to 0.73, indicating that the abstracts encapsulate the core content conveyed in the titles. Additionally, there’s a moderate to high similarity between summaries and titles, with scores ranging from 0.79 to 0.81, implying that the summaries generally reflect the main content outlined in the titles. Overall, the T5 model exhibits consistent performance across domains in aligning summaries with abstracts and titles, indicating its ability to generate coherent and relevant summaries. Further examination could delve into potential domain-specific nuances and areas for improvement to enhance summarization quality in specific topics.
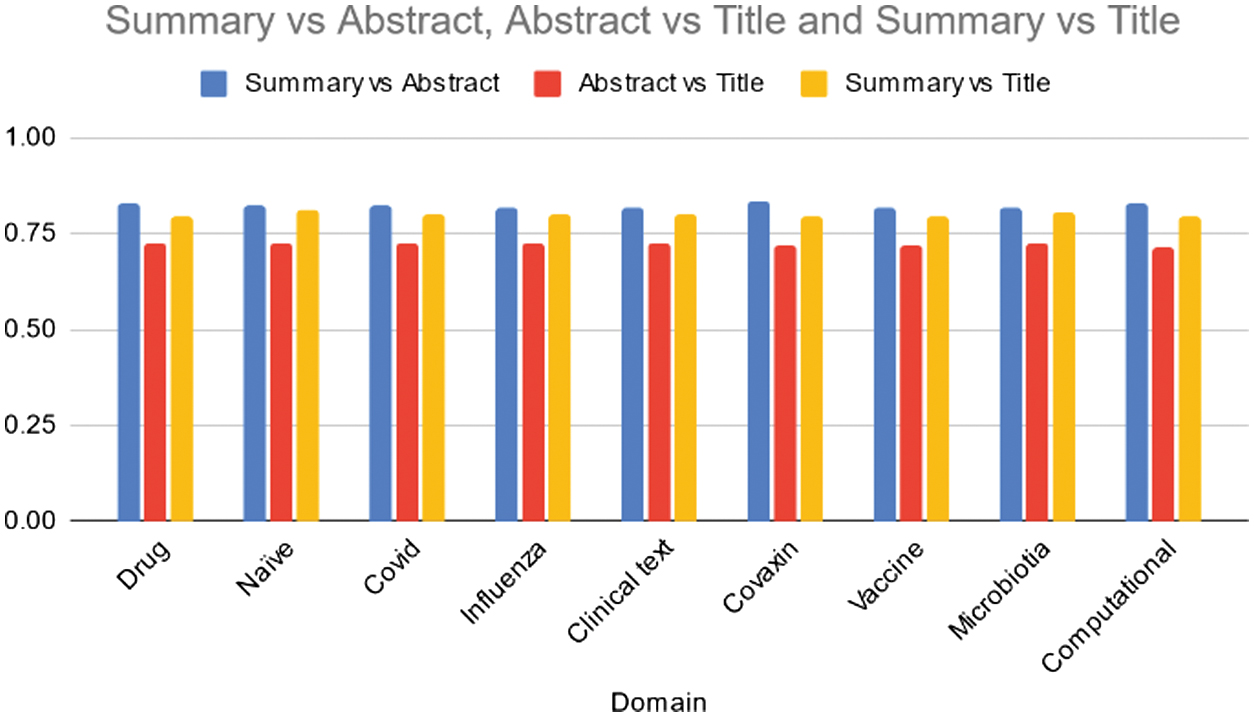 Fig. 6. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using T5 model.
Fig. 6. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using T5 model.
Table V. Average cosine similarity of the summary vs abstract, abstract vs title, and summary vs title across various domains using T5 model
| Domain | Summary vs abstract | Abstract vs title | Summary vs title |
|---|---|---|---|
| Drug | 0.83 | 0.73 | 0.79 |
| Naïve | 0.82 | 0.72 | 0.81 |
| Covid | 0.82 | 0.72 | 0.80 |
| Influenza | 0.82 | 0.72 | 0.80 |
| Clinical text | 0.82 | 0.72 | 0.80 |
| Covaxin | 0.83 | 0.72 | 0.79 |
| Vaccine | 0.82 | 0.72 | 0.80 |
| Microbiotia | 0.82 | 0.73 | 0.81 |
| Computational | 0.83 | 0.71 | 0.80 |
C.RESULTS USING TEXRANK
Table VI and Fig. 7 illustrate the ROUGE-1, ROUGE-2, and ROUGE-L scores achieved by the Texrank model across various domains. Overall, the Texrank model demonstrates moderate performance, with ROUGE-1 scores ranging from 0.14 to 0.17, ROUGE-2 scores ranging from 0.16 to 0.20, and ROUGE-L scores ranging from 0.76 to 0.82. Notably, the “Clinical text” domain exhibits the highest ROUGE-L score, indicating better fluency and coherence compared to other domains. However, the Texrank model’s performance appears to be relatively consistent across different topics, with no significant outliers. While the Texrank model performs adequately in capturing unigrams, bigrams, and longest common subsequences, there may be room for improvement in certain domains to enhance summarization quality further.
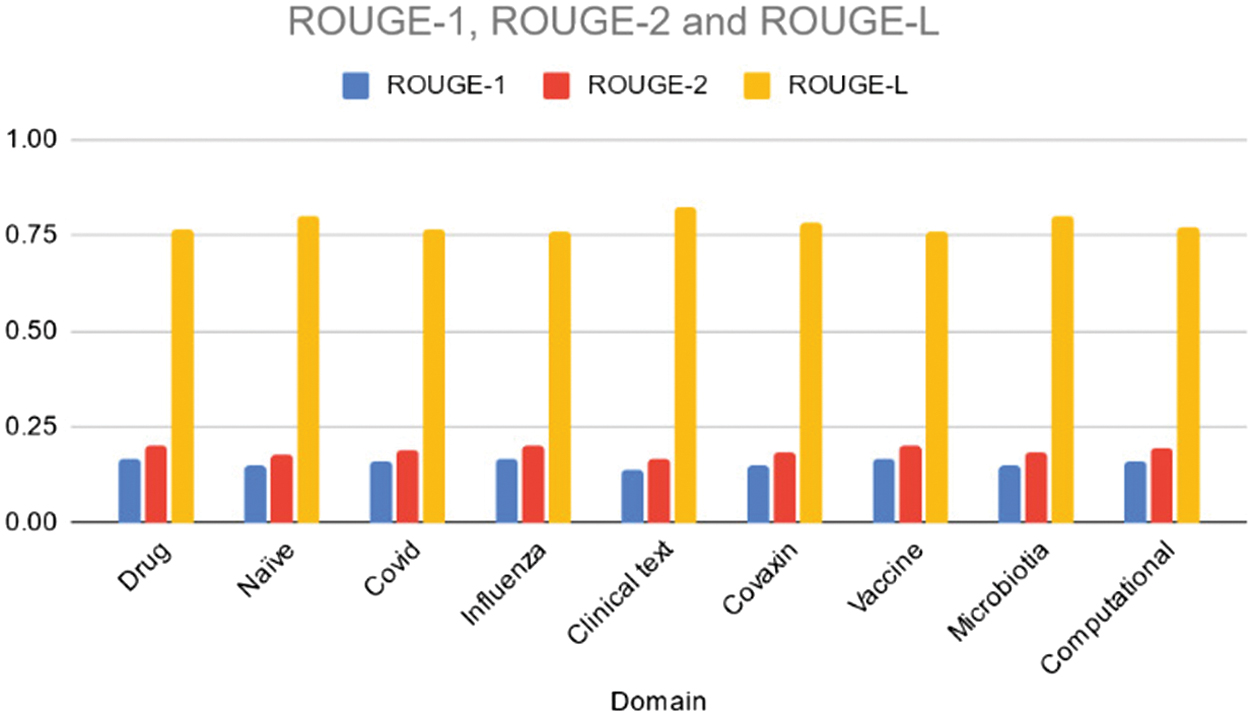 Fig. 7. Rouge 1/2/L scores of the various domains using TexRank.
Fig. 7. Rouge 1/2/L scores of the various domains using TexRank.
Table VI. ROUGE scores of various domains using TexRank model
| Domain | ROUGE-1 | ROUGE-2 | ROUGE-L |
|---|---|---|---|
| Drug | 0.17 | 0.20 | 0.77 |
| Naïve | 0.15 | 0.18 | 0.80 |
| Covid | 0.16 | 0.19 | 0.77 |
| Influenza | 0.17 | 0.20 | 0.76 |
| Clinical text | 0.14 | 0.16 | 0.82 |
| Covaxin | 0.15 | 0.18 | 0.78 |
| Vaccine | 0.16 | 0.20 | 0.76 |
| Microbiotia | 0.15 | 0.18 | 0.80 |
| Computational | 0.16 | 0.20 | 0.77 |
Table VII and Fig. 8 present cosine similarity scores obtained for data summaries using the Texrank model across various domains. Across all domains, the Texrank model consistently achieves high similarity scores between summaries and abstracts, ranging from 0.85 to 0.88.
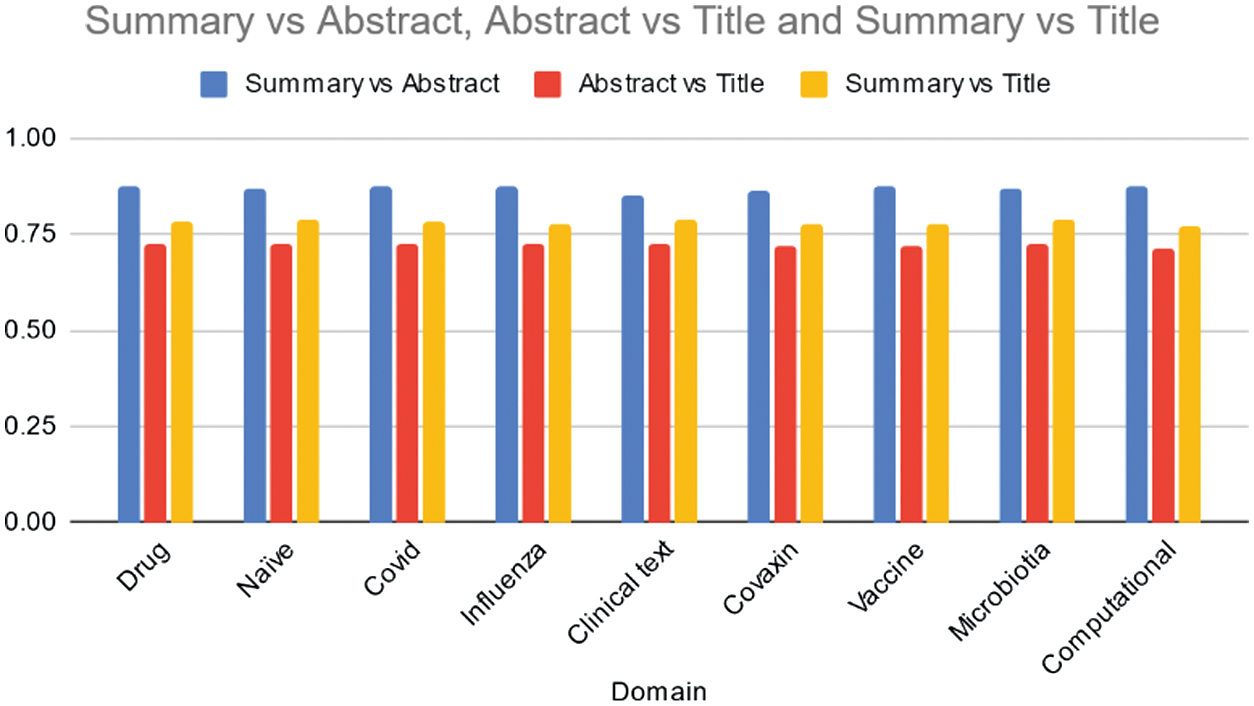 Fig. 8. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using TexRank model.
Fig. 8. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using TexRank model.
Table VII. Average cosine similarity of the summary vs abstract, abstract vs title, and summary vs title across various domains using TexRank model
| Domain | Summary vs abstract | Abstract vs title | Summary vs title |
|---|---|---|---|
| Drug | 0.87 | 0.72 | 0.78 |
| Naïve | 0.87 | 0.72 | 0.79 |
| Covid | 0.87 | 0.73 | 0.79 |
| Influenza | 0.88 | 0.73 | 0.78 |
| Clinical text | 0.85 | 0.72 | 0.79 |
| Covaxin | 0.86 | 0.72 | 0.78 |
| Vaccine | 0.88 | 0.72 | 0.77 |
| Microbiotia | 0.87 | 0.73 | 0.79 |
| Computational | 0.87 | 0.71 | 0.77 |
This suggests that the summaries generated by the model closely align with the content covered in the abstracts of the documents. Similarly, the model demonstrates strong similarity between abstracts and titles, with scores ranging from 0.71 to 0.73. This indicates that the abstracts effectively capture the key information presented in the titles of the documents. Additionally, the Texrank model shows moderate to high similarity scores between summaries and titles, ranging from 0.77 to 0.79. This suggests that the summaries generally encompass the main content conveyed in the titles of the documents, although with slightly lower consistency compared to summaries and abstracts. Table VII, Fig. 8, presents cosine similarity scores obtained for data summaries using the Texrank model across various domains. Across all domains, the Texrank model consistently achieves high similarity scores between summaries and abstracts, ranging from 0.85 to 0.88. This suggests that the summaries generated by the model closely align with the content covered in the abstracts of the documents. Similarly, the model demonstrates strong similarity between abstracts and titles, with scores ranging from 0.71 to 0.73. This indicates that the abstracts effectively capture the key information presented in the titles of the documents. Additionally, the Texrank model shows moderate to high similarity scores between summaries and titles, ranging from 0.77 to 0.79. This suggests that the summaries generally encompass the main content conveyed in the titles of the documents, although with slightly lower consistency compared to summaries and abstracts. Overall, the Texrank model exhibits consistent performance across domains, effectively capturing and summarizing the key information present in the abstracts and titles of the documents. Further analysis could explore specific areas where the model excels or requires improvement, particularly in domains where the similarity scores vary.
D.RESULTS USING LEXRANK
Table VIII and Fig. 9 present the ROUGE-1, ROUGE-2, and ROUGE-L scores achieved by the Lexrank model across different domains. Overall, the Lexrank model exhibits consistent performance, with ROUGE-1 scores ranging from 0.23 to 0.24, ROUGE-2 scores ranging from 0.30 to 0.31, and ROUGE-L scores ranging from 0.50 to 0.52. Notably, the “Microbiotia” domain shows a slightly lower ROUGE-1 score compared to other domains, indicating potential challenges in capturing unigrams effectively in this topic area.
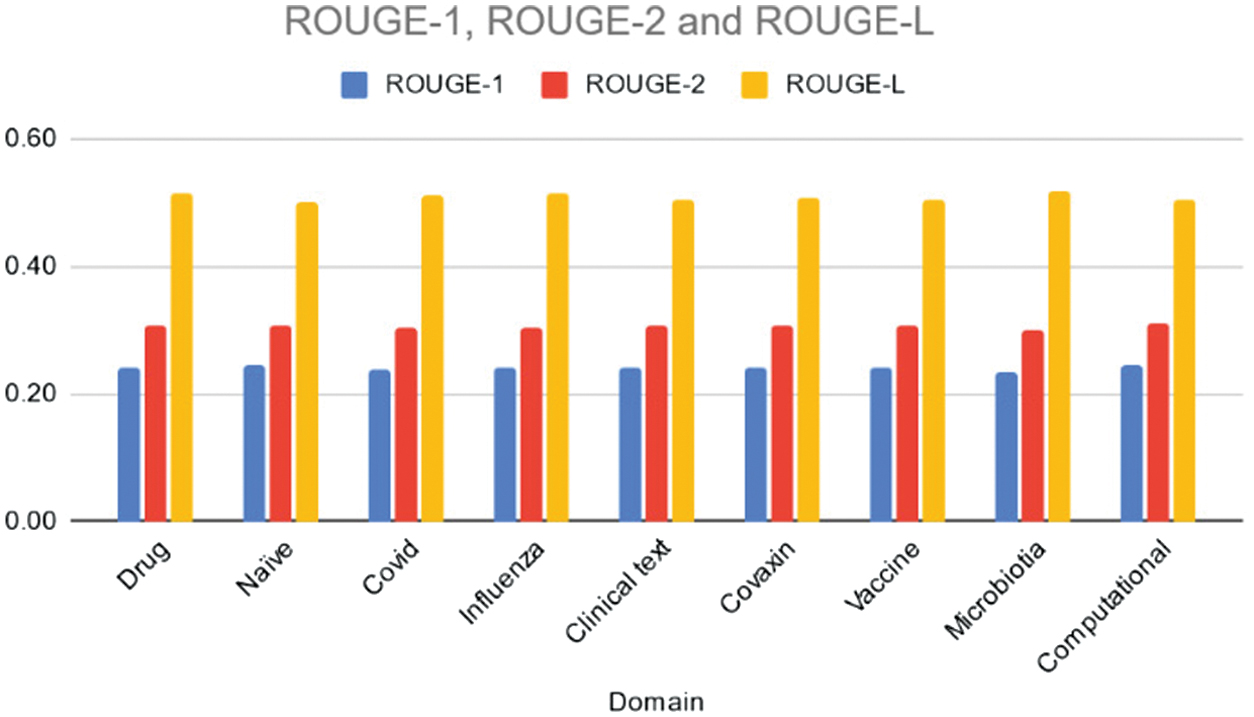 Fig. 9. Rouge 1/2/L scores of the various domains using LexRank model.
Fig. 9. Rouge 1/2/L scores of the various domains using LexRank model.
Table VIII. ROUGE scores of various domains using LexRank model
| Domain | ROUGE-1 | ROUGE-2 | ROUGE-L |
|---|---|---|---|
| Drug | 0.24 | 0.31 | 0.51 |
| Naïve | 0.24 | 0.31 | 0.50 |
| Covid | 0.24 | 0.30 | 0.51 |
| Influenza | 0.24 | 0.30 | 0.51 |
| Clinical text | 0.24 | 0.31 | 0.50 |
| Covaxin | 0.24 | 0.31 | 0.51 |
| Vaccine | 0.24 | 0.31 | 0.50 |
| Microbiotia | 0.23 | 0.30 | 0.52 |
| Computational | 0.24 | 0.31 | 0.51 |
However, the Lexrank model demonstrates relatively stable performance in capturing bigrams and longest common subsequences. While the model’s performance is consistent, further analysis could explore potential domain-specific optimizations to enhance summarization quality in specific topics. Table IX, Fig. 10 provides cosine similarity scores obtained for data summaries using the Lexrank model across various domains. Across all domains, the Lexrank model consistently achieves high similarity scores between summaries and abstracts, ranging from 0.91 to 0.92. This indicates that the summaries generated by the model closely resemble the content covered in the abstracts of the documents. Similarly, the model also demonstrates strong similarity between abstracts and titles, with scores ranging from 0.71 to 0.73. This suggests that the abstracts effectively capture the key information presented in the titles of the documents. Additionally, the Lexrank model shows moderate to high similarity scores between summaries and titles, ranging from 0.74 to 0.76.
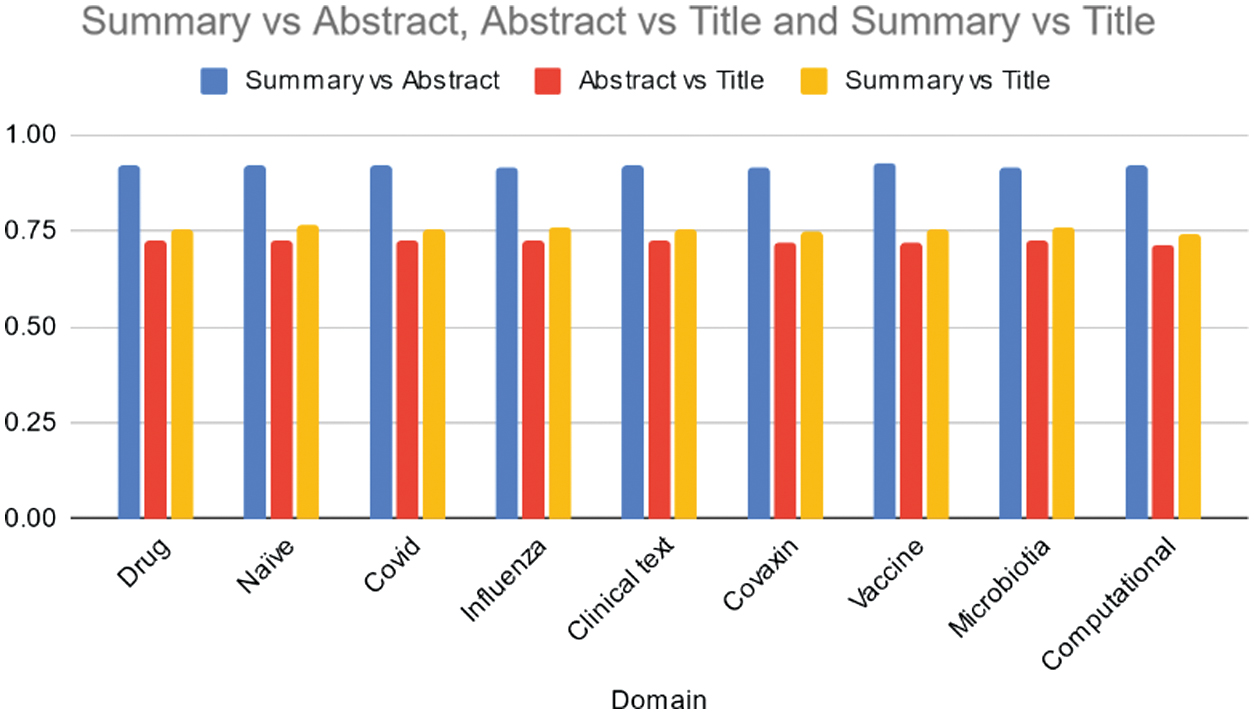 Fig. 10. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using LexRank model.
Fig. 10. Average cosine similarity of the Summary vs Abstract, Abstract vs Title, and Summary vs Title across various domains using LexRank model.
Table IX. Average cosine similarity of the summary vs abstract, abstract vs title, and summary vs title across various domains using LexRank model
| Domain | Summary vs abstract | Abstract vs title | Summary vs title |
|---|---|---|---|
| Drug | 0.92 | 0.72 | 0.75 |
| Naïve | 0.92 | 0.72 | 0.76 |
| Covid | 0.92 | 0.72 | 0.76 |
| Influenza | 0.92 | 0.73 | 0.76 |
| Clinical text | 0.92 | 0.72 | 0.75 |
| Covaxin | 0.91 | 0.72 | 0.75 |
| Vaccine | 0.92 | 0.72 | 0.75 |
| Microbiotia | 0.91 | 0.73 | 0.76 |
| Computational | 0.92 | 0.71 | 0.74 |
This indicates that the summaries generally encompass the main content conveyed in the titles of the documents, although with slightly lower consistency compared to summaries and abstracts. Overall, the Lexrank model exhibits consistent performance across domains, effectively capturing and summarizing the key information present in the abstracts and titles of the documents. Further analysis could delve into specific areas where the model may excel or require improvement, particularly in domains where the similarity scores vary.
E.DISCUSSION ABOUT THE PERFORMANCE OF VARIOUS MODELS
In summary, the comparative analysis across different summarization models – BART, T5, TexRank, and LexRank – reveals valuable insights into their performance across various domains. BART demonstrates consistent and moderate effectiveness in capturing unigrams, bigrams, and longest common subsequences, with particularly noteworthy performance in the “Clinical text” domain. Its ability to maintain strong similarity between summaries and abstracts, as well as abstracts and titles, underscores its capability to generate coherent and relevant summaries. T5, while exhibiting lower ROUGE scores compared to BART, still demonstrates reasonable performance, albeit with less consistency across domains. Despite challenges in capturing unigrams and bigrams, T5 maintains relatively high similarity scores between summaries and abstracts, suggesting its potential for generating informative summaries.
On the other hand, TexRank and LexRank models exhibit moderate to stable performance across domains. TexRank demonstrates consistent similarity between summaries and abstracts, indicating its capacity to effectively capture the essence of the information presented in the source documents. Similarly, LexRank achieves high similarity scores between summaries and abstracts, reflecting its ability to accurately summarize the content covered in the abstracts. However, it is essential to note that all models exhibit variations in performance across different domains, suggesting the need for further investigation into domain-specific optimizations to enhance summarization quality.
Overall, this comparative analysis sheds light on the strengths and weaknesses of each summarization model, offering valuable insights for researchers and practitioners in the field. While BART showcases promising performance, particularly in domains like clinical text, T5, TexRank, and LexRank also present viable options for summarization tasks. Further research into fine-tuning these models and exploring domain-specific nuances could lead to significant advancements in the field of text summarization, ultimately improving the accessibility and usability of summarized information across various domains.
F.DISCUSSION OF RESULTS ON REAL-WORLD DATASETS
We extended our analysis to include the performance of LexRank and TextRank, two popular graph-based unsupervised summarization techniques, alongside the fine-tuned transformer models (BART and T5). The comparison was conducted on both the non-medical CNN/DailyMail dataset and the medical-focused PubMed dataset, shown in Table X. The CNN/DailyMail dataset [27] is a comprehensive resource for text summarization, containing a large collection of news articles paired with multi-sentence summaries. With over 300,000 samples, it covers a wide range of topics from daily news, making it a standard benchmark for evaluating both extractive and abstractive summarization models. Its diverse content and large size provide a robust testbed for general summarization tasks.
Table X. Comparison of performance on various datasets
| Dataset | Model | ROUGE-1 | ROUGE-2 | ROUGE-L |
|---|---|---|---|---|
| CNN/DailyMail | BART | 67.5 | 48.72 | 60 |
| CNN/DailyMail | T5 | 57.53 | 45.07 | 54.79 |
| CNN/DailyMail | LexRank | 35.59 | 22.41 | 28.81 |
| CNN/DailyMail | TextRank | 35.59 | 22.41 | 28.81 |
| PubMed | BART | 21.09 | 1.37 | 10.2 |
| PubMed | T5 | 22.15 | 2.09 | 11.07 |
| PubMed | LexRank | 25.48 | 2.56 | 16.56 |
| PubMed | TextRank | 35 | 11.17 | 17.78 |
The PubMed dataset [28] consists of abstracts from biomedical literature, primarily drawn from the PubMed database. It includes over 200,000 document–summary pairs, where the summaries typically represent conclusions or key findings. This dataset is particularly challenging due to the specialized language and complex sentence structures found in medical and scientific texts, making it suitable for assessing summarization models in the context of technical and domain-specific content.
The consolidated results reveal distinct differences in model performance across the CNN/DailyMail and PubMed datasets, highlighting the strengths and limitations of various text summarization techniques. BART demonstrates superior performance on the CNN/DailyMail dataset, excelling in capturing key terms and maintaining the overall structure of general news articles, as evidenced by its high ROUGE scores. T5 also performs well on this dataset, albeit slightly below BART, showcasing its capability in general text summarization. However, both BART and T5 encounter challenges when applied to the PubMed dataset, with significantly lower scores, indicating difficulty in handling the specialized and complex language of medical texts. In contrast, LexRank and TextRank, which rely on traditional graph-based methods, perform moderately on the CNN/DailyMail dataset but surpass the transformer models on the PubMed dataset, particularly in ROUGE-1 and ROUGE-2 scores. This suggests that while transformer models are powerful for general contexts, traditional methods may be more effective in domain-specific summarization tasks, where they can better leverage the inherent structure of the text. Overall, these findings emphasize the importance of choosing the right summarization model based on the dataset’s characteristics, with transformers being more suited for general texts and traditional methods holding an edge in specialized domains like medicine.
V.CONCLUSION
In conclusion, text representation plays a crucial role in the effectiveness of text summarization techniques. While summarization using pretrained transformer models has shown promise, its application in medical and drug discovery domains remains underexplored. This study addressed this gap by focusing on extractive summarization using fine-tuned transformers and enhancing sentence representation. The task of exploring extractive text summarization in medical and drug discovery fields has been challenging due to limited datasets. To tackle this challenge, we collected abstracts from PubMed spanning various domains such as drug research and COVID-19, amounting to 1370 abstracts. Through detailed experimentation utilizing BART, T5, LexRank, and TexRank, we analyzed the dataset to perform extractive text summarization. Our findings shed light on the feasibility and efficacy of employing transformer-based models for summarizing medical and drug discovery literature, paving the way for further advancements in this area. Moving forward, there are several avenues for future research based on the findings of this study. First, exploring larger and more diverse datasets in the medical and drug discovery domains could provide deeper insights into the performance of extractive text summarization techniques using fine-tuned transformers. Additionally, investigating the applicability of other transformer models beyond BART, T5, LexRank, and TexRank may uncover novel approaches for summarizing complex medical literature. Furthermore, integrating abstractive summarization techniques into the framework could enhance the generation of concise and informative summaries, potentially capturing more nuanced information from the source texts. This could involve experimenting with transformer-based language generation models such as GPT series. Moreover, conducting user studies or evaluations involving domain experts could validate the utility and effectiveness of the generated summaries in real-world scenarios, ensuring their practical relevance and usability in aiding medical professionals and researchers. Lastly, exploring interdisciplinary collaborations with experts from both the NLP and medical domains could lead to the development of specialized tools and resources tailored to the unique challenges of medical and drug discovery text summarization, ultimately advancing the state-of-the-art in this field.