I.INTRODUCTION
Colon cancer analysis is paramount in addressing the pressing need for effective early detection and treatment strategies in combating this prevalent and deadly disease. With colon cancer ranking among the leading causes of cancer-related mortality worldwide, there is an urgent demand for robust analytical approaches to sift through vast datasets comprising patient demographics, genetic information, tumor characteristics, and treatment outcomes. These analyses aim to uncover intricate patterns and associations crucial for predicting susceptibility to the disease, prognosis, and response to therapy. By harnessing advanced analytical tools such as machine learning (ML) algorithms, researchers strive to revolutionize colon cancer management, facilitating timely interventions, personalized treatment plans, and ultimately improving patient outcomes.
High dimensionality in data mining is a significant concept, particularly in the context of colon cancer research. In this domain, datasets often contain a vast number of features, such as gene expressions, genetic mutations, and various biomarkers. These high-dimensional datasets present both challenges and opportunities for effective data analysis and mining [1].
One primary challenge of high dimensionality in colon cancer research is the “curse of dimensionality.” As the number of features increases, the data space becomes exponentially larger, leading to sparsity. This sparsity complicates the identification of meaningful patterns and can render traditional data mining techniques less effective or computationally impractical. High-dimensional data can also result in overfitting of predictive models, where the model captures noise instead of underlying biological patterns, leading to poor performance on new, unseen data [2].
To overcome these challenges, researchers employ dimensionality reduction techniques. Methods like principal component analysis and linear discriminant analysis are used to transform the high-dimensional data into a lower-dimensional space, preserving as much relevant information as possible. Feature selection techniques are also crucial, as they help in identifying and retaining the most significant features that contribute to understanding colon cancer while reducing the dimensionality of the dataset [3].
Imbalanced datasets are a common issue in data mining and ML, particularly in the context of colon cancer research. An imbalanced dataset occurs when the classes or categories in the data are not represented equally [4]. For instance, in colon cancer studies, the number of samples from patients with cancer (positive class) might be significantly lower than those from healthy individuals (negative class). This imbalance poses unique challenges for data analysis and model training such as biased models, evaluation metrics, and overfitting [5]. This article majorly focuses on the addressing of high dimensionality and imbalanced issues of colon cancer using ML approaches.
ML algorithms are becoming increasingly popular across various fields beyond colon cancer. In healthcare, ML techniques are used for lung cancer classification [6], enabling more accurate and early detection of the disease. In cybersecurity, ML is employed to detect phishing attacks by identifying patterns and anomalies in email and web data that suggest fraudulent activities [7]. In education, ML enhances personalized learning experiences, predicting student performance and tailoring educational content to individual needs [8]. Additionally, in demographic studies, ML is applied to gender classification, analyzing facial features, voice patterns, and other attributes to determine gender [9]. These diverse applications highlight the versatility and power of ML in solving complex problems, improving efficiency, and providing valuable insights across different domains. As the technology continues to advance, its integration into various sectors is expected to grow, driving innovation and better outcomes in each field.
The need for colon cancer prediction using ML algorithms stems from the disease’s significant public health impact and the potential to improve patient outcomes through early detection and personalized treatment strategies [10]. Colon cancer is one of the most prevalent and deadly forms of cancer worldwide, with high mortality rates, particularly when diagnosed at advanced stages [11]. Early detection is critical for successful treatment, yet many cases are not identified until symptoms appear or through routine screening, which may occur too late for effective intervention. ML algorithms offer the promise of more accurate and timely prediction by leveraging complex patterns within large datasets, enabling healthcare professionals to identify individuals at high risk of developing colon cancer and tailor screening and prevention strategies accordingly [12]. Additionally, these algorithms can help optimize treatment plans by predicting patient responses to different therapies, facilitating more personalized and effective care approaches.
ML and data mining are closely related fields that involve extracting insights and patterns from data. While they share some similarities, they also have distinct focuses and methodologies. ML and data mining involve several stages: problem definition, data collection, preprocessing, exploratory data analysis (EDA), feature selection/engineering, model selection, training, evaluation, tuning, deployment, and monitoring/maintenance. In problem definition, objectives and criteria are established, followed by data collection from various sources. Preprocessing cleans and transforms the data, and EDA explores it for insights. Feature selection/engineering enhances relevant variables, and model selection involves choosing appropriate algorithms. Models are then trained on data, evaluated for performance, and tuned as needed. Finally, successful models are deployed into production, with ongoing monitoring and maintenance to ensure continued effectiveness [13].
As discussed above, preprocessing is a very important phase and consumes 80 to 90% of total data analysis. In preprocessing, class imbalance and feature selection play a very important role in achieving better results [14]. In this research also we addressed the imbalance and feature selection issues for the prediction of colon cancer. Let us see the importance and its implications of both of those.
A.IMBALANCING
Class imbalance in ML refers to situations where the distribution of classes in the dataset is heavily skewed, with one class significantly outnumbering the others. This imbalance can lead to biased models that favor the majority class, resulting in poor performance for the minority class [15]. The implications of class imbalance include reduced predictive accuracy, inflated evaluation metrics for the majority class, and difficulty in identifying rare but important patterns [16]. Moreover, models trained on imbalanced data may struggle to generalize well to new data, leading to decreased overall performance. Addressing class imbalance is crucial, often requiring techniques such as resampling methods (e.g., oversampling and undersampling), algorithmic adjustments (e.g., class weights), or using evaluation metrics that are robust to imbalance (e.g., F1 score and area under the precision-recall curve). Failure to account for class imbalance can result in biased and ineffective models, impacting the reliability and usefulness of ML systems [17]. The various researchers presented methods for handling class-imbalanced data to improve classification performance [18,19].
Class imbalance in medical datasets can have significant implications for ML models used in healthcare applications. In such datasets, it is common to encounter scenarios where certain medical conditions are relatively rare compared to others. For instance, in a dataset of patients, the number of individuals with a particular disease might be significantly smaller than those without it. This class imbalance can lead to several challenges. First, models trained on imbalanced medical data may exhibit a bias toward the majority class, potentially resulting in false negatives for the minority class, i.e., failing to correctly identify patients with the rare condition. This can have serious consequences in healthcare, where early detection and accurate diagnosis are critical. Additionally, imbalanced medical datasets can skew evaluation metrics, making it appear as though a model is performing well when it is actually failing to detect important medical conditions. Addressing class imbalance in medical datasets is therefore paramount, requiring careful consideration of sampling techniques, algorithmic adjustments, and the choice of evaluation metrics to ensure the reliability and effectiveness of ML models deployed in healthcare settings.
B.HIGH DIMENSIONALITY
High dimensionality in ML refers to datasets with a large number of features or variables, which can present challenges in model training and performance. The importance of feature selection lies in mitigating these challenges by identifying and retaining only the most relevant features that contribute meaningfully to the predictive task. High dimensionality can lead to increased computational complexity, overfitting, and decreased generalization performance of ML models. Feature selection helps address these issues by reducing the dimensionality of the data, thereby improving model efficiency, interpretability, and predictive accuracy [20]. By selecting the most informative features, feature selection also aids in enhancing model robustness, reducing noise, and facilitating better insights into the underlying relationships within the data, ultimately leading to more effective and reliable ML models.
In medical datasets, high dimensionality can have significant implications for ML applications. With numerous features representing various medical attributes, such as patient demographics, symptoms, lab results, imaging data, and genetic markers, high-dimensional datasets pose challenges for modeling and analysis [21]. The abundance of features can lead to increased computational complexity during training, making it computationally expensive and time-consuming to process and analyze the data. Furthermore, high dimensionality can exacerbate the risk of overfitting, where models learn noise or irrelevant patterns from the data, potentially resulting in poor generalization performance on unseen data.
Feature selection becomes crucial in this context as it helps mitigate these challenges by identifying the most informative features relevant to the medical prediction or classification task. By reducing the dimensionality of the dataset through feature selection, models can achieve better generalization, interpretability, and performance, leading to more accurate and clinically relevant predictions or diagnoses. Moreover, feature selection aids in extracting meaningful insights from complex medical data, facilitating improved decision making by healthcare professionals and potentially enhancing patient outcomes.
The rest of the article is organized in three more sections. In which Section II details the literature review of class imbalance and high dimensionality for prediction of colon cancer and similar diseases with different algorithms. Research methodology is elaborated in Section III. Experimental result analysis is articulated in Section IV. Finally, the article is concluded with possible recommendations in Section V.
II.LITERATURE REVIEW
In the literature, several researchers contributed different techniques to predict colon cancer. In this section. some of those are discussed.
The researchers [22] discussed the use of tumor aggression score (TAS) as a prognostic factor for determining the tumor stages of colon cancer, highlighting its importance and sensitivity in benefiting the TNM staging. The top-performing model, random forest, achieved an accuracy of 0.84 and an AUC of 0.82 ± 0.10 for predicting the five years disease-free survival (DFS) of the colon cancer patients. Additionally, it was observed that patients with TAS ≥9.8 had poor DFS, while those with TAS <9.8 had a DFS exceeding 10 years.
The approach outlined in the article [23] involves utilizing a comprehensive dataset of colorectal cancer patient information to develop predictive models using various ML techniques, including random forest, general linear model, and neural network algorithms. These models were trained to predict clinically relevant outcomes, with particular emphasis on dichotomous outcomes such as relapse and RCT-R. Impressively, the most successful models achieved accuracies of 0.71 and 0.70 for relapse and RCT-R, respectively, when evaluated on blinded test data. Additionally, the prediction models for overall survival and DFS demonstrated strong performance, as evidenced by C-Index scores of 0.86 and 0.76, respectively.
The article [24] presented a model for individualized survivability prediction for colon cancer patients over five years after treatment, using a classification approach and ML techniques, based on the SEER dataset. It aims to determine the ideal number of features for prediction and operationalize the prediction model in an application. The article also focuses on developing a system that can accept specific inputs and produce outputs for each year of survival after treatment. The performance of the six-feature model is close to that of the model using 18 features.
Koppad et al. [25] introduced a novel approach utilizing random forest methodology to explore CRC gene associations through ML. Their methodology encompassed six distinct ML methods employed as classifiers, with the use of the GridSearchCV function to determine optimal values for each model’s hyperparameters. To assess performance, the analysis was conducted across three GEO datasets, utilizing a combinatorial approach with the six ML models for training and testing data comparison. The study incorporated multiple validation strategies to ensure robustness and reliability of the model’s performance evaluation.
The gradient boosting model has the largest area under the Receiver Operating Characteristics curve 0.82 [26]. The methodology involved utilizing a specific cancer dataset for training and testing ML algorithms, using eight different algorithms, reporting experiment results, and evaluating the importance of top risk factors.
The random forest approach is recommended when modeling high-dimensional microarray data [27]. The methodology involved using a dataset of gene expression levels, applying statistical analysis and the Synthetic Minority Oversampling Technique (SMOTE) method to address class imbalance, using the LASSO feature selection method to select 13 genes associated with colon cancer, and employing random forest, decision tree, and Gaussian naive Bayes methods for modeling. In the current research also class imbalance is addressed using SMOTE. But employed the different classifiers along with different feature selection methods.
The study described in reference [28] focuses on developing prognostic prediction models for the survival time of colon cancer patients’ post-surgery, emphasizing the significance of high-risk molecular features combined with specific clinical characteristics. Factors such as age over 70, T3 stage, poorly differentiated or undifferentiated tumors, M0 stage with well-differentiated tumors, M0 stage with T2 tumors, high lymph node ratio (LNR), and T4 stage with poorly differentiated or undifferentiated tumors are highlighted as critical for 5-year survival. The research underscores the importance of early diagnosis and the identification of predictive biomarkers in improving patient outcomes. Employing ML and statistical methods, the study utilizes data from SEER and TCGA databases to construct these predictive models, with a particular emphasis on the significant role of the positive LNR in the prognosis model.
The proposed methodology achieved a classification accuracy of up to 94.36% in distinguishing colorectal cancer patients from normal individuals using a combination of Z-normalization, Fisher score for gene selection, K-means clustering for representative gene selection, and the modified harmony search algorithm for feature selection [29]. The feature selection method using the harmony search algorithm led to high classification accuracy by using only a few genes, with an artificial neural network (ANN) as the classifier. The ANN used in the study had input and hidden layers composed of five nodes, an output layer consisting of one node, and utilized the sigmoid function as the activation function.
The study achieved a promising 98.4% accuracy for cancer classification after feature selection [30]. The feature selection method significantly improved the classification accuracy from 95.2% to 98.4% on the colon cancer dataset [31]. The results of the experiment were comparable with other studies on colon cancer research, indicating a significant improvement and promising future applications.
After the review, it has been observed that ranking-based feature selections, subset feature selections are not tested for the prediction of colon cancer. This current research addresses those missing components and analyzes its result before and after addressing the class imbalance problem as the dataset collected is not balanced. Those details are discussed in subsequent sections.
III.METHODOLOGY
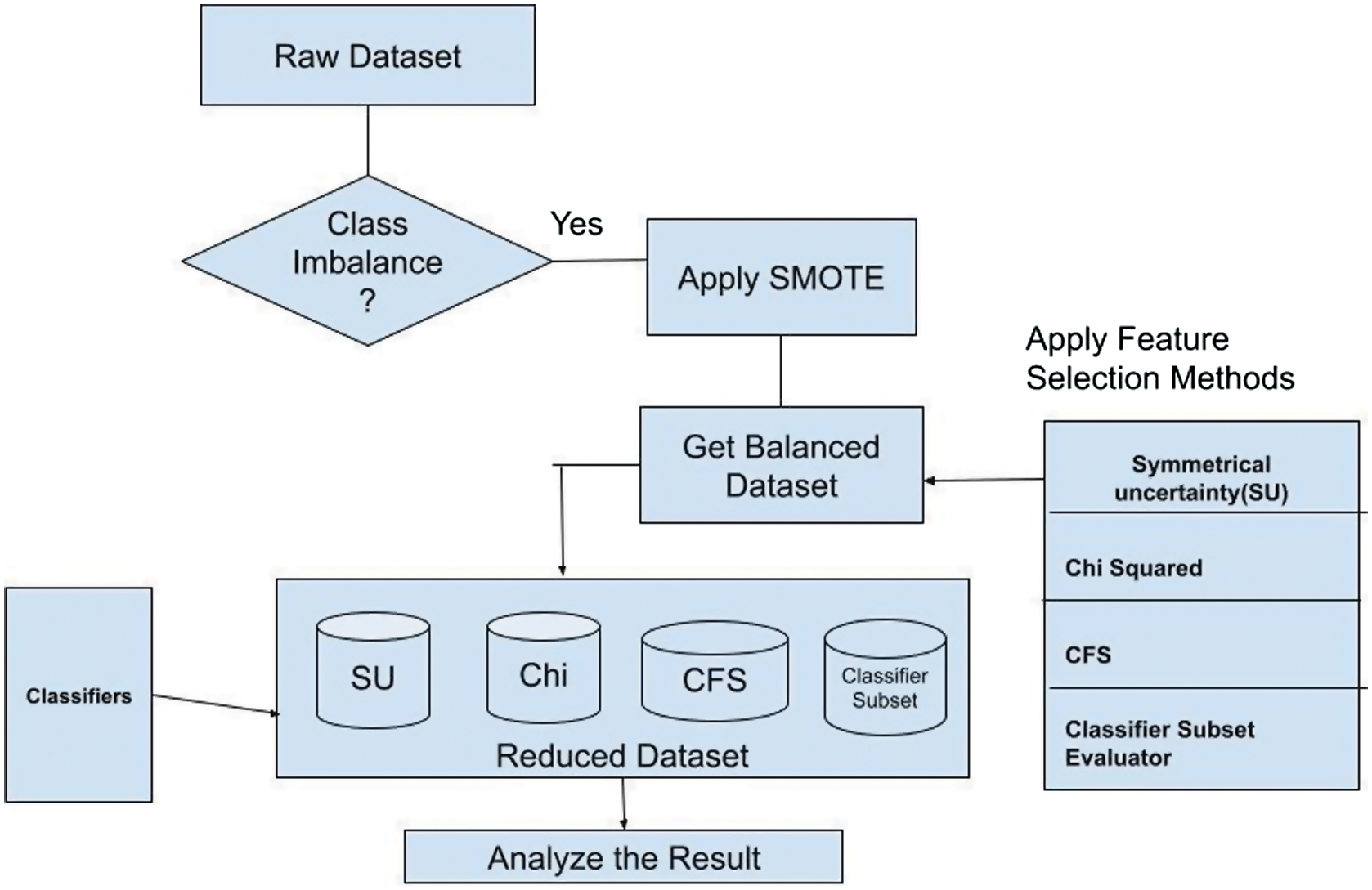
The proposed methodology is based on the three factors that includes test class imbalance, if the dataset is imbalanced, resolve it with SMOTE. Apply the filter-based ranking methods and subset evaluators to select the best features over the both balanced and imbalance then apply the classifier set as mentioned in the research architecture.
As per the Fig. 1, initially raw dataset is first collected. Later class imbalance is tested. The collected dataset is imbalanced, i.e., data points are not distributed properly according to the class labels. If this would be the case, the learning algorithm will be biased toward the majority instance class. So, a popular technique called SMOTE is applied on the imbalanced dataset to make the balance. SMOTE is based on the K-NN algorithm, it will add synthetic instances to the existing dataset. So that dataset will become balanced.
As the dataset has a huge number of features (2000), in order to minimize the training time and increase the accuracy, strong features are selected using various statistical filter-based ranking feature selection methods such as symmetrical uncertainty (SU) and Chi-square attribute evaluator (Chi). These are based on the concept of information theory in which a score is assigned to each feature based on the values that those features contain. Depending on the problem statement, we can choose the top ranked features. In addition, two wrapper approach feature selection methods such as correlation-based feature selection (CFS) subset evaluator and classifier subset evaluator are also applied to get the subset of features.
SU is a measure of the amount of uncertainty shared between two variables. It is commonly used in feature selection to evaluate the relevance of features in a dataset. The higher the SU value between a feature and the target variable, the more relevant that feature is for predicting the target.
The formula for symmetrical uncertainty between two variables X and Y is given by
Here:- •IG(X,Y) is the information gain between variables X and Y.
- •H(X) and
- •H(Y) are the entropies of variables X and Y, respectively.
The SU value ranges from 0 to 1. A value of 0 indicates no mutual information between the variables (i.e., no feature relevance), while a value of 1 indicates complete mutual information (i.e., perfect feature relevance). In feature selection, one would typically calculate the SU values for each feature with respect to the target variable and select the features with the highest SU values as the most relevant features for the predictive task.
The Chi-square (χ2) test is a statistical method used to determine the independence of two categorical variables. In the context of feature selection, the Chi-square test is commonly employed to evaluate the relevance of features by measuring the dependency between a categorical feature and a categorical target variable.
The formula for the Chi-square statistic between a feature X and a target variable Y is given by
Oi is the observed frequency of each category in feature X.Ei is the expected frequency of each category in feature X, which is calculated under the assumption of independence between X and Y.
The Chi-square statistic is calculated by summing the squared differences between observed and expected frequencies, normalized by the expected frequencies. A higher Chi-square value indicates a stronger association between the feature and the target variable.
The CFS subset evaluator is a method used for feature selection that aims to select features that are highly correlated with the target variable but uncorrelated with each other. This helps in identifying a subset of features that collectively provide predictive power while avoiding redundancy.
The formula for CFS evaluates the “merit” of a subset of features by considering both the individual feature relevance (measured through the correlation with the target variable) and the redundancy among the features in the subset.
The merit of a subset S is calculated as follows:
where- •k is the number of features in the subset S.
- •avgCor(S,C) is the average correlation of features in S with the target variable C.
- •avgRed(S) is the average pairwise correlation between features in S.
The CFS subset evaluator aims to maximize the merit value, indicating a high correlation with the target variable while minimizing redundancy among the selected features. It achieves this by searching for subsets of features that strike the right balance between relevance and redundancy.
The classifier subset evaluator is a feature selection method that evaluates the usefulness of a subset of features based on their performance in classification tasks. It works by training a classifier on different subsets of features and measuring their performance using some evaluation metric, such as accuracy, F1-score, or area under the receiver operating characteristic curve (area under the curve).
To test the strength of the features selected by various methods and to show that the balanced dataset will produce better accuracy. Different classifiers such as rule based, tree based, and lazy learners are applied on both balanced and imbalanced datasets.
Here is the brief description of the classifiers considered. J48 is an implementation of the C4.5 algorithm, which is used to generate a decision tree. Decision trees classify data by splitting it into subsets based on feature values, forming a tree-like model of decisions. It handles both categorical and continuous data, performs pruning to avoid overfitting, and is easy to interpret. Simple cart (classification and regression trees) is an algorithm that builds binary decision trees. It recursively splits the data into two groups to maximize the homogeneity within each group. It can handle both classification and regression tasks, is robust to outliers, and provides a clear visual representation of decision-making.
Jrip is an implementation of the Repeated Incremental Pruning to Produce Error Reduction (RIPPER) algorithm, which generates a set of if-then rules for classification. It is efficient for large datasets, performs rule pruning to reduce complexity, and can handle both binary and multi-class classification. Ridor stands for RIpple-DOwn Rule learner, which generates a default rule and then exceptions to this rule to handle classification. It focuses on reducing error rates by creating exceptions to general rules, and it is effective for datasets with many attributes.
K nearest neighbor (KNN) is a lazy learning algorithm that classifies data points based on the k-nearest training examples in the feature space. It makes decisions based on the majority class among the neighbors. It is simple and intuitive, nonparametric, and effective for small to medium-sized datasets. However, it can be computationally intensive for large datasets. SVM is a powerful supervised learning algorithm that finds the optimal hyperplane to separate different classes in the feature space. It works well for both linear and nonlinear data by using kernel functions. It provides high accuracy, is effective in high-dimensional spaces, and works well with a clear margin of separation between classes. It is particularly effective for binary classification tasks.
IV.EXPERIMENT AND RESULT
As discussed in the research methodology in the previous section, the experiment is conducted to show the impact of high dimensionality and imbalancing of dataset for prediction of Colon Cancer [28]. The colon cancer data utilized in this study were obtained from publicly available medical databases, which include comprehensive records of patient information, diagnostic results, and treatment outcomes. These databases ensure the reliability and quality of the data, enabling accurate analysis and reproducibility of the research findings [32]. The initial dataset has 2000 instances, 2 class labels (tumor and normal), and 62 instances as shown in Fig. 2. Out of which 40 belongs to tumor and 22 are normal. This distribution shows the imbalancing. So, the dataset is balanced by applying SMOTE as described in the research methodology.
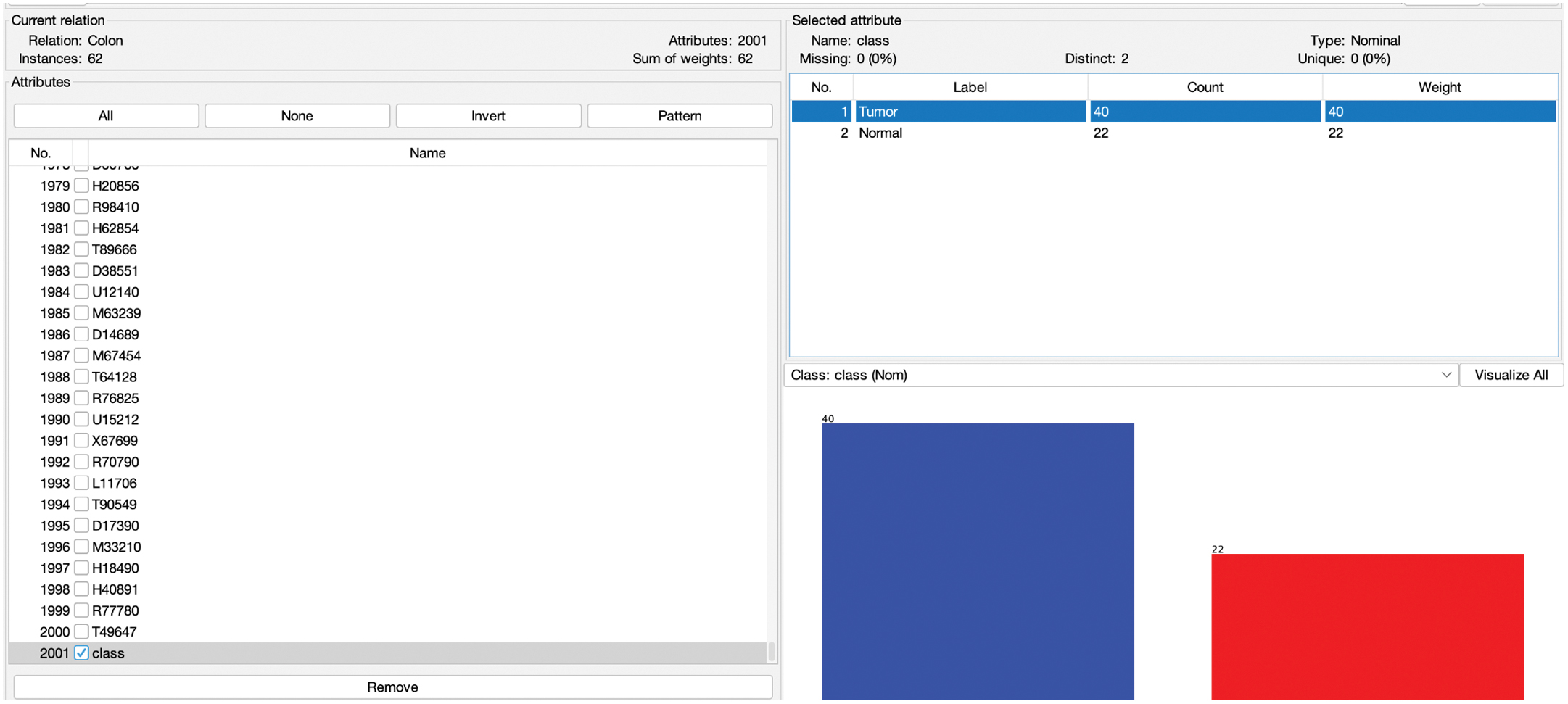 Fig. 2. Distribution of imbalanced dataset.
Fig. 2. Distribution of imbalanced dataset.
After applying SMOTE for the normal classed instance with 100% rate. 22 more synthetics instances are created. With this effect, the dataset now has 84 instances in which 40 belong to tumor class and 44 normal class, thereby minimizing the imbalancing ratio. As the dataset has 2000 features in it. To reduce the dimensionality, several feature selection methods as described in the methodology section are applied on both the datasets, i.e., balanced and imbalanced. SMOTE was utilized to address data imbalancing. However, a potential limitation of SMOTE is the risk of overfitting due to the creation of synthetic instances that may not fully capture the underlying data distribution. To mitigate this, we combined SMOTE with careful feature selection and cross-validation techniques. By selecting only the most relevant features and validating the model performance on different subsets of the data, we aimed to reduce the likelihood of overfitting and ensure the robustness of the predictive models.
First, SU is applied on an imbalanced dataset; it derived 135 features with a score above 0. As the feature score less than 0 cannot contribute anything to the learning model, remaining features are ignored for the training. Same thing happened with the Chi-squared evaluator also. The correlation-based feature selection (CFS) has produced only a strong subset with 25 features and classifier subset evaluator derived 205 features.
The same features selection methods are applied to the balanced dataset. In that case, SU and Chi derived 456 features, CFS produced 34 features, and classifier subset evaluator derived 205 features.
To test the strength of those features both with balanced and imbalanced datasets various classifiers such as Jrip and Ridor (rule-based classifiers), J48 and simple cart (tree-based classifiers), K nearest neighbor (lazy learner), and support vector machine (SVM) are applied and those performances are recorded. In addition to this feature selection, it was tested with a full dataset also. The whole experiment was conducted with the help of a popular WEKA tool. Table I shows the classification accuracy of each learner with respect to the feature selection techniques on an imbalanced dataset. The same is demonstrated in the graphical way in Fig. 3.
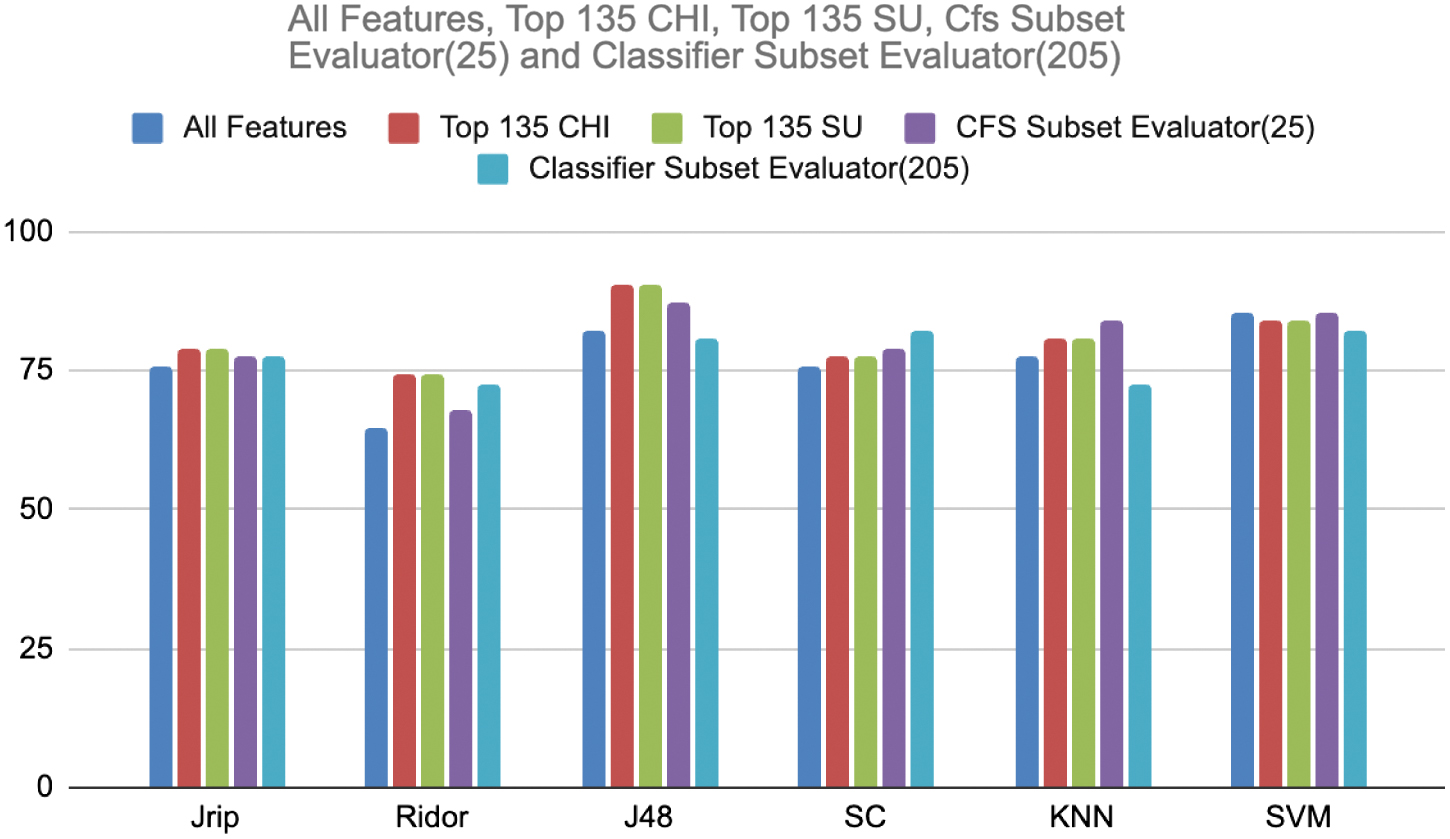 Fig. 3. Comparison of classification accuracy on imbalanced dataset.
Fig. 3. Comparison of classification accuracy on imbalanced dataset.
TABLE I. Classification accuracy on imbalanced dataset
| Jrip | Ridor | J48 | SC | KNN | SVM | |
|---|---|---|---|---|---|---|
| 79.03 | 74.19 | 90.32 | 77.41 | 80.64 | 83.87 | |
| 79.03 | 74.19 | 90.32 | 77.41 | 80.64 | 83.87 | |
| 77.41 | 67.74 | 87.09 | 79.03 | 83.87 | 85.48 | |
| 77.41 | 72.58 | 80.64 | 82.25 | 72.58 | 82.25 |
Above result analysis revealing the power of the feature selection. When all the features are considered, Jrip recorded 75.8% accuracy, whereas all feature selection methods achieved better than it. Chi and SU secured high accuracy, i.e., 79.03%. The same is true with Ridor and J48 also except the classifier subset evaluator in case of J48. Ridor recorded 74.19 with Chi and SU. J48 produced 90.32 with SU and Chi which is highest. With the classifier subset evaluator SC recorded 82.25 which is higher than all feature selections. With CFS, subset evaluator KNN and SVM marked the higher accuracy 83.87% and 85.48%, respectively, than all feature selection methods.
Table III displays the classification accuracy of each learner with respect to the feature selection techniques on a balanced dataset. The same is demonstrated in the graphical way in Fig. 4.
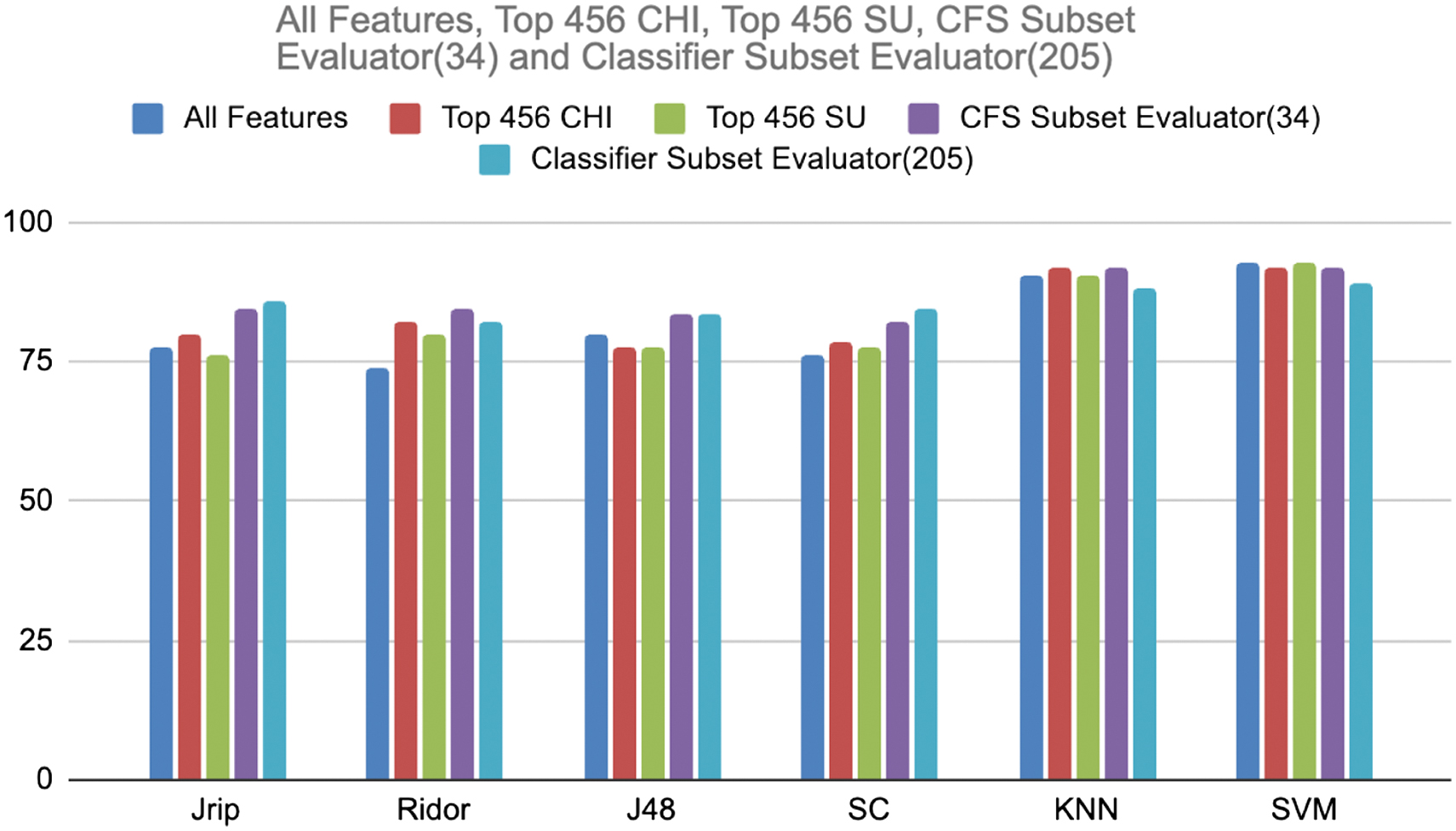 Fig. 4. Comparison of classification accuracy on imbalanced dataset.
Fig. 4. Comparison of classification accuracy on imbalanced dataset.
As per Table II statistics, classifier subset evaluator produced 85.71% higher accuracy than all feature selectors with Jrip. All the feature selectors performed better with Ridor, out of which CFS recorded higher accuracy, i.e., 84.52%. With J48, CFS, and classifier subset evaluator marked the 83.33% accuracy. With SC, classifier subset evaluator marked the 84.52% accuracy. With KNN classifiers, CHI and CFS recorded 91.66% higher accuracy. SVM is not performed well with feature selectors. The same comparison shown in Fig. 3 in a graphical way.
TABLE II. Classification accuracy on imbalanced dataset
| Jrip | Ridor | J48 | SC | KNN | SVM | |
|---|---|---|---|---|---|---|
| 77.38 | 73.8 | 79.76 | 76.19 | 90.47 | 92.85 | |
| 79.76 | 82.14 | 77.38 | 78.57 | 91.66 | 91.66 | |
| 76.19 | 79.76 | 77.38 | 77.38 | 90.47 | 92.85 | |
| 84.52 | 84.52 | 83.33 | 82.14 | 91.66 | 91.66 | |
| 85.71 | 82.14 | 83.33 | 84.52 | 88.09 | 89.28 |
TABLE III. Comparison of classification accuracy on balanced and imbalanced dataset with Jrip, Ridor, J48
| Jrip | Ridor | J48 | ||||
|---|---|---|---|---|---|---|
| Imbalanced | Balanced | Imbalanced | Balanced | Imbalanced | Balanced | |
| 77.38 | 73.8 | 79.76 | ||||
| 79.03 | 79.76 | 74.19 | 82.14 | 90.32 | 77.38 | |
| 79.03 | 76.19 | 74.19 | 79.76 | 90.32 | 77.38 | |
| 77.41 | 84.52 | 67.74 | 84.52 | 87.09 | 83.33 | |
| 77.41 | 85.71 | 72.58 | 82.14 | 80.64 | 83.33 | |
Classifier and feature selection wise compilation articulated in Tables III and IV on both balanced and imbalanced datasets.
Table IV. Comparison of classification accuracy on balanced and imbalanced dataset with SC, KNN, SVM
| SC | KNN | SVM | ||||
|---|---|---|---|---|---|---|
| Imbalanced | Balanced | Imbalanced | Balanced | Imbalanced | Balanced | |
| 76.19 | 90.47 | 92.85 | ||||
| 77.41 | 78.57 | 80.64 | 91.66 | 83.87 | 91.66 | |
| 77.41 | 77.38 | 80.64 | 90.47 | 83.87 | 92.85 | |
| 79.03 | 82.14 | 83.87 | 91.66 | 85.48 | 91.66 | |
| 82.25 | 84.52 | 72.58 | 88.09 | 82.25 | 89.28 | |
Tables III and IV reveals that most of the feature selectors perform better on the balanced dataset. Thus, it is recommended to balance the dataset then remove the duplicate or irrelevant features before applying the learning techniques.
V.CONCLUSION
In this study, we investigated the critical aspects of imbalanced data and feature selection in the context of colon cancer prediction. Our findings underscored the significance of preprocessing techniques in enhancing the performance of predictive algorithms. By addressing the imbalance issue and employing various feature selection methods, including Chi-square, symmetrical uncertainty, CFS subset, and classifier subset evaluators, we observed notable improvements in the accuracy of popular classifiers such as tree, rule lazy (KNN), and SVM. The results demonstrated that after balancing the dataset and applying appropriate feature selection methods, all algorithms exhibited enhanced performance. Notably, KNN achieved a remarkable accuracy of 91.66% with Chi-square and CFS, while SVM attained an impressive accuracy of 92.85% with symmetrical uncertainty. Other classifiers, such as Jrip, Rider, J48, and Simple Cart, also showed significant improvements in accuracy across different feature selection methods.
These findings emphasized the importance of preprocessing steps, such as balancing imbalanced data and selecting relevant features, in optimizing predictive models for colon cancer detection. The proposed research framework offers valuable insights into improving prediction accuracy, thereby contributing to advancements in early detection, treatment outcomes, public health initiatives, research efforts, and overall patient care related to colon cancer. Future research could further explore additional preprocessing techniques and evaluate their impact on predictive performance in diverse datasets and clinical settings. Future research could explore hybrid approaches combining oversampling and undersampling techniques to achieve a more balanced dataset while minimizing the risk of overfitting and information loss. Additionally, investigating advanced feature selection methods and integrating deep learning models could further enhance the predictive accuracy and generalizability of colon cancer detection systems.