I.INTRODUCTION
Citrus fruits, such as grapes, oranges, lemons, and limes, are among the most widely cultivated and commercially important fruit crops globally. Their nutritional composition is highly regarded for its abundance of vitamin C, flavonoids, antioxidants, and dietary fiber. Its versatility in the fields of industry, medicine, and gastronomy adds to its worth. Individuals worldwide consume and utilize a variety of products derived from citrus fruits, including juices, marmalades, essential oils, and dietary supplements. The citrus business generates billions of dollars annually and contributes significantly to the agricultural economy, supporting employment, international trade, food security, and rural lifestyles.
Despite extensive cultivation and numerous benefits, several plant diseases are progressively undermining the quantity and quality of citrus crops. Among the most common and detrimental are those that particularly impact citrus foliage. Foliar diseases are a significant concern, as leaves are essential for photosynthesis and are often the first places to show signs of infection. These preliminary signs are crucial for the prompt diagnosis and treatment of diseases. Poorly managed or ignored issues can have significant adverse effects on farmers’ profitability by lowering fruit output, quality, market value, and esthetic appeal.
Citrus leaves are typically impacted by diseases such as citrus canker, greasy spot, Huanglongbing (HLB), citrus black spot, and citrus tristeza virus (CTV). Each of these diseases exhibits unique signs such as elevated lesions, discoloration, chlorosis, mottling, curling, vein clearing, and premature leaf abscission. The bacterium Xanthomonas citri causes citrus canker, characterized by elevated, corky lesions encircled by yellow halos. Greasy spot is induced by the fungus Mycosphaerella citri, presenting as oily lesions on the undersides of leaves, ultimately resulting in defoliation. The dots exhibit a yellowish-brown coloration. HLB, the most detrimental leaf disease, is induced by a species of Candidatus Liberibacter and is characterized by irregular, blotchy mottling. The fungal disease known as citrus black spot, or Phyllosticta citricarpa, adversely affects leaves and fruit by producing resilient, necrotic lesions. CTV, a viral affliction, simultaneously manifests chlorosis, stem pitting, vein clearing, and an overall decline in tree vitality. The effective treatment of these symptoms relies on prompt and precise identification, given their severity and complexity. An infographic delineates the symptoms of the predominant diseases affecting citrus leaves. Fig. 1 illustrates symptoms of major citrus leaf diseases.
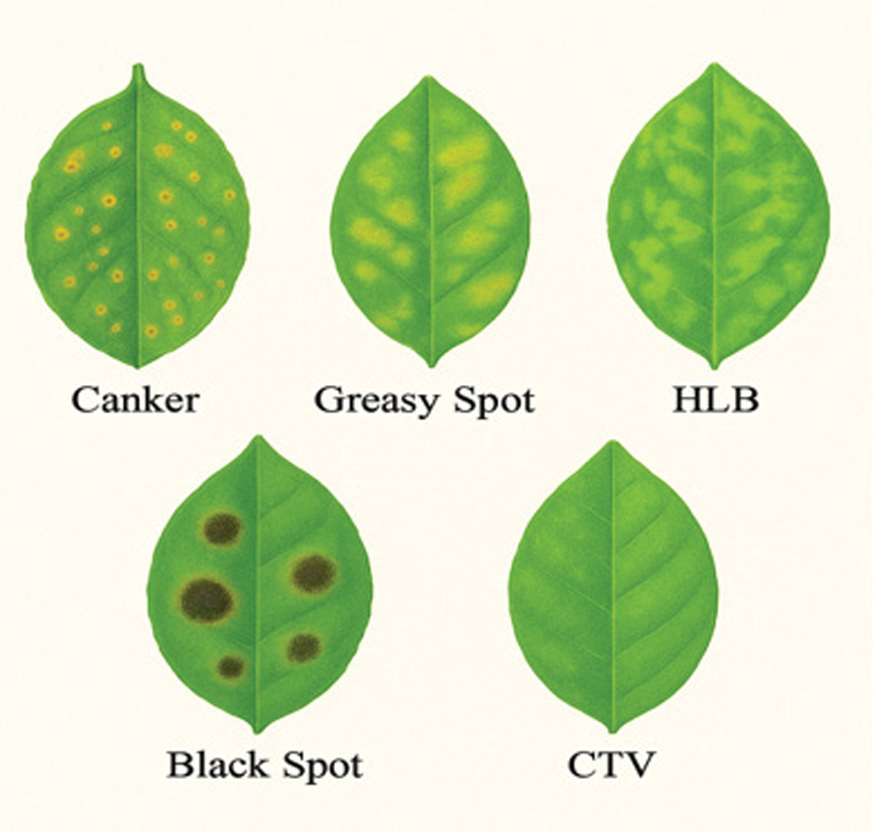 Fig. 1. Representative symptoms of major citrus leaf diseases (canker, greasy spot, HLB, black spot, and CTV).
Fig. 1. Representative symptoms of major citrus leaf diseases (canker, greasy spot, HLB, black spot, and CTV).
Historically, extension agents, plant pathologists, and agronomists have conducted first-hand examinations of citrus leaves to diagnose illnesses. While efficient in regulated environments, this manual method still possesses numerous drawbacks. Since visual diagnosis is subjective, it can be affected by the inspector’s background, the environment, and how similar symptoms are across different diseases or nutritional deficiencies. Manual methods are labor-intensive, costly, unreliable, and inefficient in areas that require close supervision or lack expert access. In situations with poor lighting or the initial phases of a disease when symptoms are mild, human error, exhaustion, and misidentification can reduce the accuracy of a diagnosis.
The increasing global demand for citrus fruits and the expansion of citrus farming into new environmental regions necessitate the urgent development of more efficient, scalable, and dependable disease detection and classification methods. Recent years have witnessed advancements in agricultural diagnostics facilitated by the integration of image analysis with artificial intelligence (AI) and deep learning technologies. Convolutional neural networks (CNNs) are among the most promising techniques in this domain. CNNs have demonstrated outstanding efficacy in numerous computer vision applications, such as object detection, facial recognition, medical imaging, and, more recently, plant disease identification.
CNNs excel at discerning visual patterns and spatial hierarchies in images through the utilization of many convolutional and pooling layers. These networks are ideal for examining the complex and varied visual characteristics of damaged plant leaves, as they can autonomously extract feature representations—such as color, texture, shape, and edge information—from raw pixel data. A critical determinant of CNN success is the accessibility of extensive, annotated datasets for model training. In agricultural settings, challenges in data collection and labeling, along with environmental and varietal discrepancies in disease manifestation, can lead to inadequate datasets.
Transfer learning has emerged as a solution to the problem of insufficient data, gaining considerable appeal. Transfer learning is the refinement of CNN models for particular tasks, such as citrus disease classification, subsequent to their pretraining on extensive, diverse picture datasets like ImageNet. This method reduces the necessity for extensive domain-specific datasets, accelerates model convergence, and enhances accuracy by leveraging the general visual attributes obtained during pretraining. Transfer learning has demonstrated notable efficacy in agricultural applications due to the scarcity of labeled data and the substantial variability of experimental circumstances compared to real-world scenarios.
While transfer learning offers benefits, it is not invariably the case that utilizing a singular CNN architecture produces the best results. Each CNN model possesses distinct architectural constraints, meticulously optimized to capture specific characteristics of image quality. While it may lack the capacity to comprehend intricate patterns, one of the initial deep learning models, AlexNet, is efficient and cost-effective in computation. Despite introducing processing overhead, VGG16’s profound and uniform architecture facilitates enhanced feature discrimination. ResNet50 employs residual connections to facilitate the training of extremely deep networks and mitigate vanishing gradient issues. This enables the extraction of more complex properties. Each model possesses distinct advantages and disadvantages, underscoring the shortcomings of current disease classification methods. Table I provides a comparative overview of the attributes and architectures of various CNNs.
Table I. Comparison of ensemble models with proposed model for citrus disease classification
| Model | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| MMFN (AlexNet + VGG16 + ResNet50) | 0.975 | 0.966 | 0.980 | 0.985 |
| Ensemble (VGG16 + ResNet50 + DenseNet121) | 0.990 | 0.985 | 0.965 | 0.966 |
| Ensemble (ResNet50 + InceptionV3 + EfficientNetB0) | 0.985 | 0.980 | 0.955 | 0.960 |
Ensemble learning techniques have gained prominence in recent years as a potential alternative to the constraints of individual models. Ensemble learning integrates the predictive capabilities of multiple models to improve overall classification performance. Ensemble approaches enhance generalizability, robustness, and variance reduction by integrating the optimal attributes of several architectures. Ensemble approaches facilitate the integration of diverse feature representations obtained from many CNNs, hence improving the accuracy and comprehensiveness of plant disease diagnosis.
This study presents a unique framework for categorization, the multi-model fusion network (MMFN), which integrates the advantages of ensemble learning with transfer learning. The MMFN utilizes the synergies of three advanced CNN architectures: ResNet50, VGG16, and AlexNet, as shown in Fig. 2. Each model is meticulously refined on a well-selected and annotated dataset of citrus leaf pictures, encompassing both healthy and diseased specimens. The outputs from each CNN are amalgamated through a weighted fusion method to prioritize the most pertinent and reliable predictions in the final categorization. This ensemble methodology enhances the sensitivity and specificity of disease detection, facilitating a more accurate differentiation between seemingly equivalent diseases. Table I illustrates the comparison of CNN architectures and the MMFN model pipeline from an architectural perspective. Table II represents a comparison of CNN architectures for citrus disease classification.
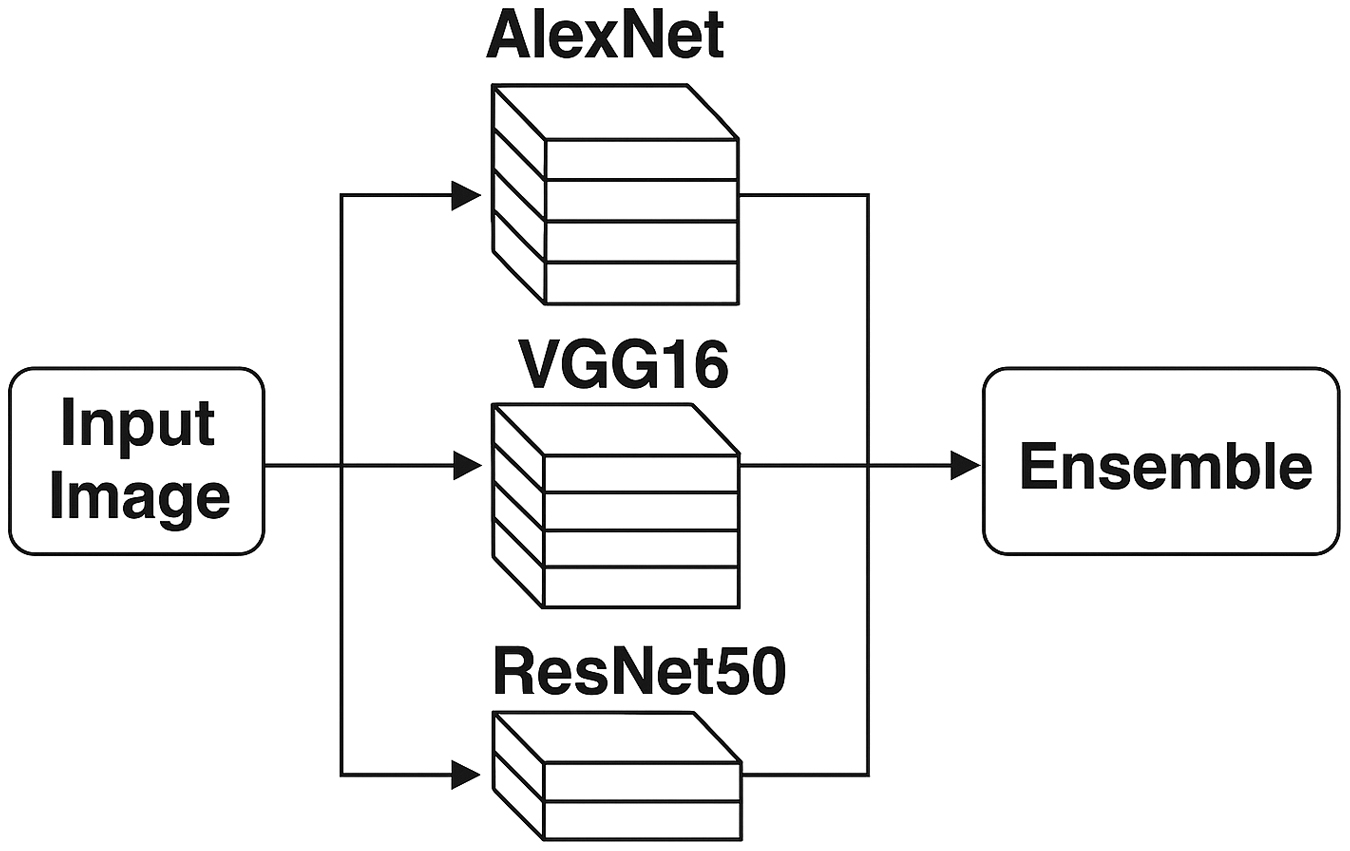 Fig. 2. MMFN architecture: multi-model ensemble pipeline using ResNet50, VGG16, and AlexNet.
Fig. 2. MMFN architecture: multi-model ensemble pipeline using ResNet50, VGG16, and AlexNet.
Table II. Dataset composition for citrus leaf disease classification
| Disease type | Number of samples | Visual characteristics |
|---|---|---|
| Citrus canker | 800 | Lesions with yellow halos |
| Greasy spot | 750 | Oily, yellowish-brown spots |
| HLB | 900 | Blotchy, asymmetric mottling |
| Citrus black spot | 700 | Round black necrotic lesions |
| Citrus tristeza virus | 850 | Stem pitting, vein clearing |
| Healthy | 1000 | Uniform green foliage |
Our diverse dataset ensures that the MMFN is resilient and adaptable to real-world agricultural environments by encompassing a wide array of variations in disease severity, leaf orientation, lighting conditions, and background clutter. Features include visual markers like halos, mottling, necrotic spots, vein clearing, and color uniformity. The model’s performance is assessed by many quantitative metrics. Included are recall, accuracy, precision, F1-score, and the analysis of the confusion matrix. These measures evaluate the model’s prediction capability and applicability in real-world agricultural settings. The classification performance of all models and the MMFN ensemble is presented in Table II.
This study aims to develop a diagnostic tool for the early detection and management of citrus leaf diseases by farmers, agronomists, and agricultural extension professionals. The tool should be intuitive, adaptable, and intelligent. The proposed system can be integrated with mobile apps, drones, and field-deployed imaging devices for immediate, on-site diagnosis of illnesses. This approach allows stakeholders to quickly apply pesticides, prune, or quarantine-affected plants, thereby improving yields, reducing crop losses, and promoting more sustainable citrus farming.
This work further expands the growing body of research on the use of modern AI technology in precision agriculture. The MMFN marks a significant advancement in developing cost-effective, accurate, and scalable solutions for plant health monitoring by combining ensemble deep learning with transfer learning. Its adaptability to various crops and diseases allows the proposed approach to support larger projects aimed at enhancing global food security and encouraging sustainable farming practices.
This study addresses three key challenges in plant disease classification: data scarcity, model inconsistency, and practical deployment. The proposed MMFN method, which combines transfer learning with ensemble CNNs and a carefully chosen dataset, will help automate disease detection in citrus leaves. This effort aims to connect advanced AI research with agricultural practices, improving diagnostic accuracy, reducing financial losses, and speeding up the global adoption of smart farming technologies.
The remainder of this paper is organized as follows. Section II reviews related literature. Section III details the proposed methodology. Section IV presents the experimental results and analysis. Finally, Section V concludes the paper with key findings and future directions.
II.LITERATURE REVIEW
A.TRADITIONAL METHODS OF CITRUS DISEASE DETECTION
Citrus fruits, among the most frequently farmed crops, are significantly affected in yield and quality by several foliar diseases. Experienced farmers and agricultural specialists typically use visual inspection to detect these issues. This manual screening method can identify indicators of disease, such as spots, discoloration, or leaf lesions. Although it has notable limitations, this approach is often employed in resource-limited, small-scale agricultural settings. Visual diagnosis can be inaccurate when symptoms are faint, early-stage, or resemble other diseases due to its subjective nature, labor intensity, and heavy dependence on human skill [1,2]. Environmental factors like lighting conditions, plant maturity, or stress levels may obscure or entirely erase disease symptoms, making precise identification difficult.
Several laboratory-based, non-visual diagnostic techniques have been studied to address these limitations. Included in this group are hyperspectral and multispectral imaging, which detect spectral signatures on leaves that might otherwise go unnoticed. These methods can identify physiological stress early in the development of a disease [3,4]. Specific techniques for diagnosing particular diseases and stress markers include serological assays, enzyme-linked immunosorbent assay (ELISA), electrochemical sensors, and fluorescence imaging [5,6].
Although their precision is high, their high cost, requirement for specialized equipment, and need for technical knowledge make them impractical for large-scale agricultural use. This suggests that most people only encounter them in research institutions or well-funded commercial farming operations. To address the accuracy issue while maintaining scalability, solutions using computer vision and digital photography have been proposed [7].
B.MACHINE LEARNING APPROACHES FOR PLANT DISEASE DETECTION
The proliferation of machine learning (ML) has revolutionized plant disease diagnosis by facilitating automated and scalable detection methods. ML algorithms may detect leaves based on disease characteristics and discern patterns from visual data. Initial plant disease research employs popular classifiers such as Support Vector Machines (SVM), Random Forest (RF), Decision Trees, and k-Nearest Neighbors (kNN) [8,9].
Generally, these methods rely on manually designed features such as edge orientation histograms, shape descriptors, texture metrics (e.g., Gray-Level Co-occurrence Matrix (GLCM), Local Binary Pattern (LBP)), and color histograms. Domain experts identify these attributes and subsequently feed them into the ML model for classification [10]. This technique, while exhibiting some success, lacks generalizability and reliability. Handmade qualities are typically task-specific and sometimes prove ineffective when applied to different plant species or novel disease types [11]. Variations in lighting, backdrop obstructions, or leaf positioning during image acquisition may adversely affect their performance in uncontrolled environments.
Moreover, traditional ML models fail to acquire hierarchical feature representations, resulting in a deficiency in the semantic understanding of intricate visual patterns that deep learning models may achieve. As a result, research focus has increasingly shifted to deep learning methodologies that autonomously extract features from raw picture data, hence eliminating the necessity for manual feature engineering [12].
C.DEEP LEARNING FOR CITRUS LEAF DISEASE CLASSIFICATION
CNNs have revolutionized image-based illness classification by their ability to automatically extract multi-level characteristics from input images. CNNs are particularly effective for identifying citrus leaf diseases due to their hierarchical architecture, which captures both low-level features (e.g., edges, corners) and high-level characteristics (e.g., lesion shapes, textures). Models such as AlexNet, VGG16, and ResNet50 have been extensively tested on citrus disease datasets and often achieve accuracy rates above 95%, significantly outperforming traditional ML techniques [14].
CNNs have the advantage of being end-to-end trainable, which simplifies the model pipeline and reduces human intervention. They also exhibit better adaptability to subtle variations in disease manifestation, such as color changes and leaf deformation, that are difficult to model using fixed feature extractors [15]. However, CNN performance often declines when exposed to domain shifts, such as changes in environmental lighting, image resolution, or background noise [16].
Additionally, CNNs require large volumes of labeled training data to generalize well. This presents a major hurdle in agriculture, where curated datasets are limited due to the cost and expertise required for accurate annotation [17]. To address this data scarcity, researchers have increasingly turned to transfer learning and ensemble learning as strategies to improve classification accuracy while maintaining model generalizability and efficiency [18].
1)TRANSFER LEARNING IN AGRICULTURAL IMAGE CLASSIFICATION
Transfer learning involves reusing pretrained models—originally developed on large datasets like ImageNet—for domain-specific tasks such as citrus disease classification. The fundamental idea is to transfer acquired traits from general image recognition to specific agricultural applications, hence reducing data and processing requirements.
In plant disease classification initiatives, fine-tuning models like ResNet, InceptionV3, DenseNet121, and MobileNetV2 consistently yield superior results. These designs permit the selective retraining of specific layers to accommodate new sickness categories while preserving acquired visual representations [21,22]. In instances with limited, imbalanced, or noisy datasets, where traditional training from scratch may lead to overfitting, transfer learning has demonstrated its advantages.
Recent advancements encompass hybrid transfer learning methodologies that amalgamate features from many pretrained models. Research has demonstrated that integrating shallow models, like AlexNet, with deeper models, such as ResNet50 or DenseNet, yields a diverse feature representation that enhances performance on complex classification tasks ([23,24]).
Moreover, transfer learning accelerates training and facilitates deployment on resource-constrained devices such as smartphones and drones, making it very suitable for real agricultural applications.
D.ENSEMBLE LEARNING FOR ROBUST CLASSIFICATION
While transfer learning enhances the performance of individual models, ensemble learning augments accuracy and robustness by integrating many classifiers. Majority voting, bagging, boosting, and stacking are ensemble strategies that amalgamate the predictions of several models to achieve a more reliable and accurate outcome [25,26].
Ensemble approaches have been utilized in citrus disease detection to amalgamate predictions from many CNN architectures. Research has demonstrated that ensembles of AlexNet, VGG16, and ResNet50 surpass individual models because of their complementary strengths in feature extraction [27]. For instance, while AlexNet excels at identifying large-scale features, VGG16 is adept at discerning fine-grained textures, and ResNet50 captures deeper semantic information.
Feature-level fusion, which involves integrating intermediate representations from many networks and transmitting them through a final classification layer, is an exceptionally effective ensemble method. This method reduces the likelihood of misclassification due to model bias and enables the model to utilize a broader feature space.
Plant phenotyping and disease detection systems are progressively utilizing deep ensemble learning as the norm. When integrated with explainability tools, such models demonstrate enhanced generality, improved interpretability, and greater resilience to noisy inputs [29,30].
1)CHALLENGES AND FUTURE RESEARCH DIRECTIONS
Despite substantial advancements in deep learning and ensemble approaches, numerous challenges continue to hinder their broader application in practical agriculture. Class imbalance in medical datasets, characterized by the predominance of common diseases and the under-representation of rare diseases, constitutes a serious issue. Consequently, models may exhibit bias toward majority classes, leading to diminished recall for minority conditions [31].
Techniques like data augmentation, synthetic data generation using Generative Adversarial Networks (GANs), and sampling strategies (e.g., SMOTE, undersampling) have been proposed to tackle this problem [32]. Moreover, the use of semi-supervised or unsupervised learning could reduce the dependency on labeled data by leveraging unlabeled images to learn meaningful representations [33].
Another major concern is the computational complexity of CNNs, particularly in ensemble settings where multiple deep models are run concurrently. This limits real-time deployment on resource-constrained devices like field sensors or smartphones [34]. Future research should prioritize lightweight models such as MobileNetV3, EfficientNet, or NASNet, which balance accuracy and efficiency.
Additionally, the integration of deep learning with Internet of Things (IoT) platforms opens new opportunities for real-time disease monitoring. Coupled with edge computing, this integration could allow farmers to detect diseases instantly using mobile applications or Unmanned Aerial Vehicle (UAV)-mounted cameras.
Lastly, explainability and transparency of AI models remain underexplored in agricultural contexts. Tools like Grad-CAM, SHAP, and LIME can offer visual insights into which regions of an image influenced a model’s prediction. These build trust among end-users and facilitate model validation by domain experts.
III.METHODOLOGY
Accurate citrus leaf disease classification is achieved using MMFN, which combines transfer learning and ensemble learning through design and execution. The approach is organized into the following main phases. Figure 3 shows the flowchart of the approach.
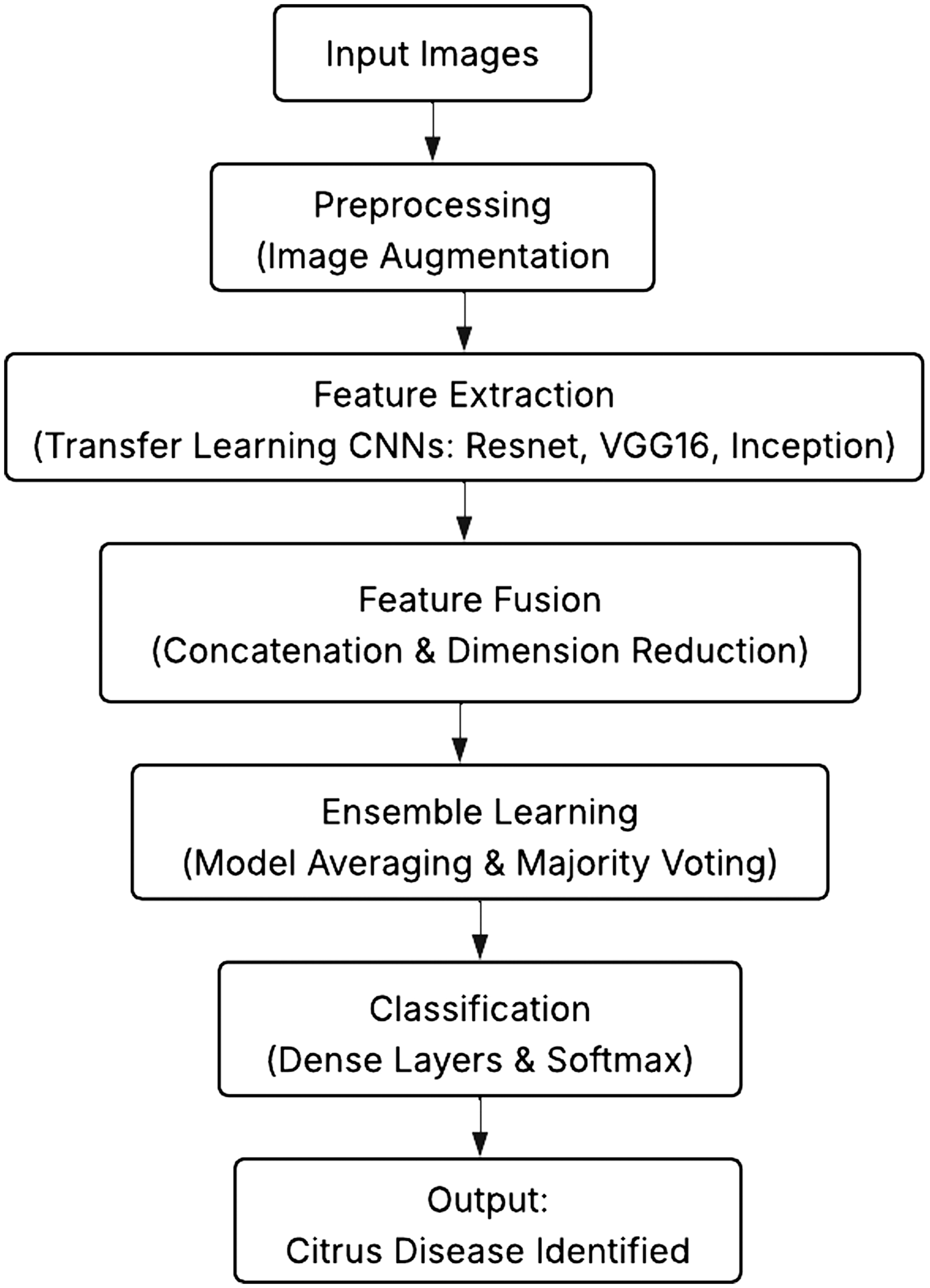 Fig. 3. Flowchart illustrating the methodology.
Fig. 3. Flowchart illustrating the methodology.
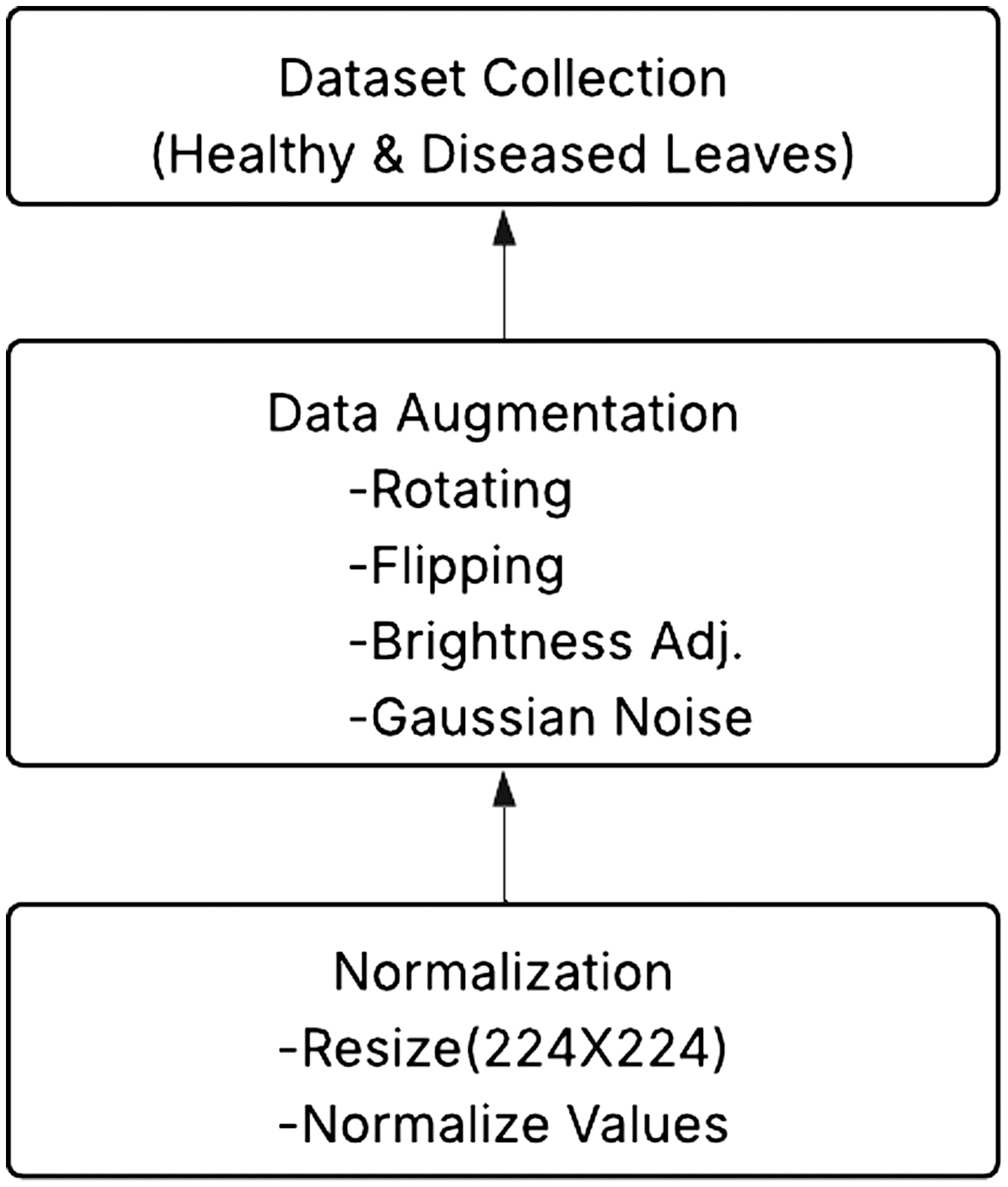
A.DATA COLLECTION AND PREPROCESSING
Public collections and field sources provide a thorough dataset of citrus leaf photos. The dataset contains pictures of healthy and sick leaves impacted by citrus canker, greasy spot, HLB, citrus black spot, and CTV.
B.DATA AUGMENTATION
The dataset undergoes augmentation methods, including rotation, flipping, brightness correction, and Gaussian noise addition, to increase generalization and address class imbalance. Images are downsized to a consistent size, for example, 224 × 224 pixels, and normalized to match the input requirements of the pretrained models. Figure 4 illustrates the preprocessing and data gathering process.
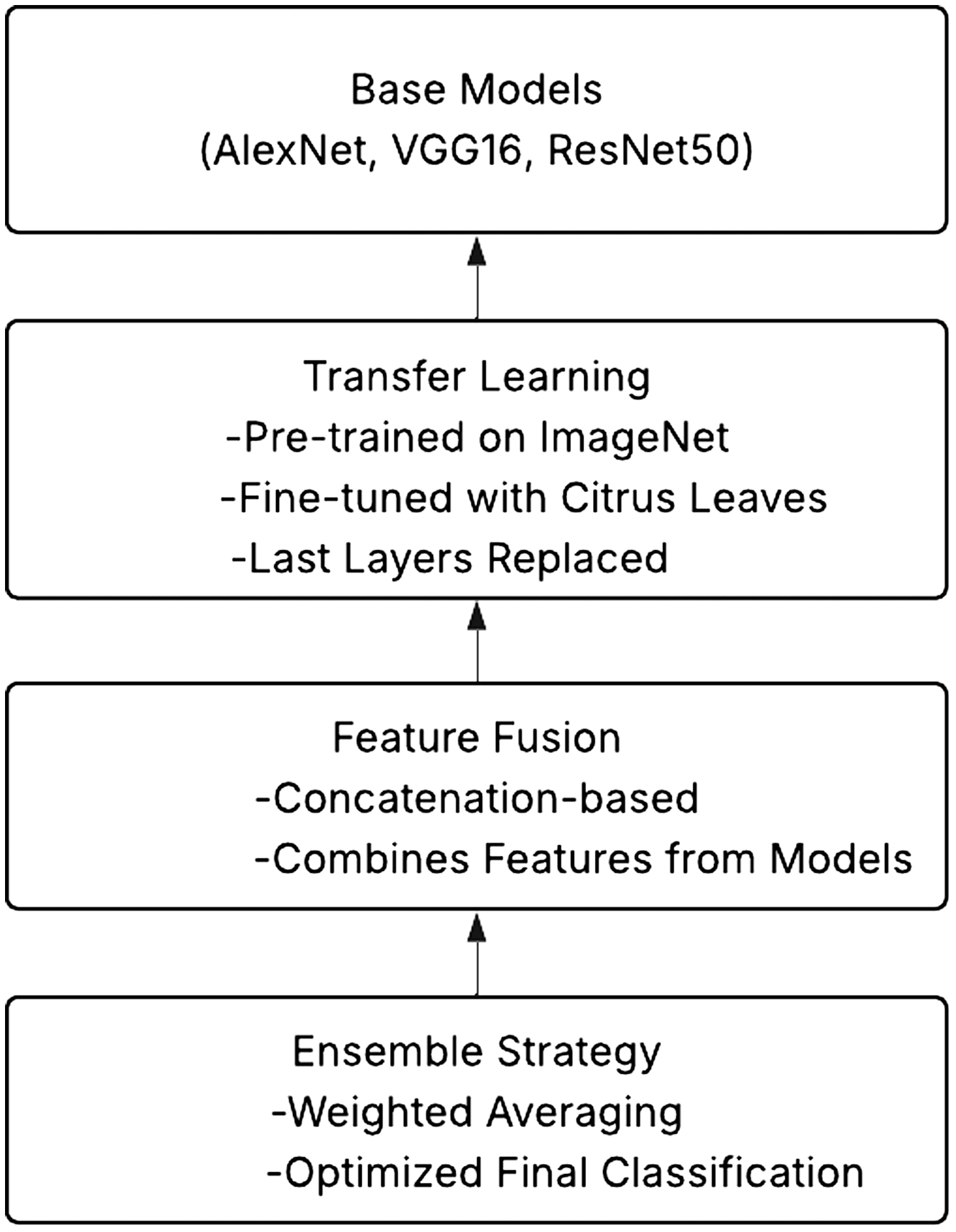
C.MULTI-MODEL FUSION NETWORK (MMFN) ARCHITECTURE
Base Models: Feature extraction through transfer learning is performed on three well-known CNN architectures—AlexNet, VGG16, and ResNet50—as shown in Fig. 5. Citrus leaf photos are used to fine-tune pretrained ImageNet models. Fully connected layers replace the last layers, enabling classification into healthy and sick categories.
Feature Fusion: A concatenation-based feature fusion method combines features from the three models to enhance representation learning. Using a weighted average ensemble approach, the final classification is performed, thereby maximizing performance by leveraging the strengths of each model.
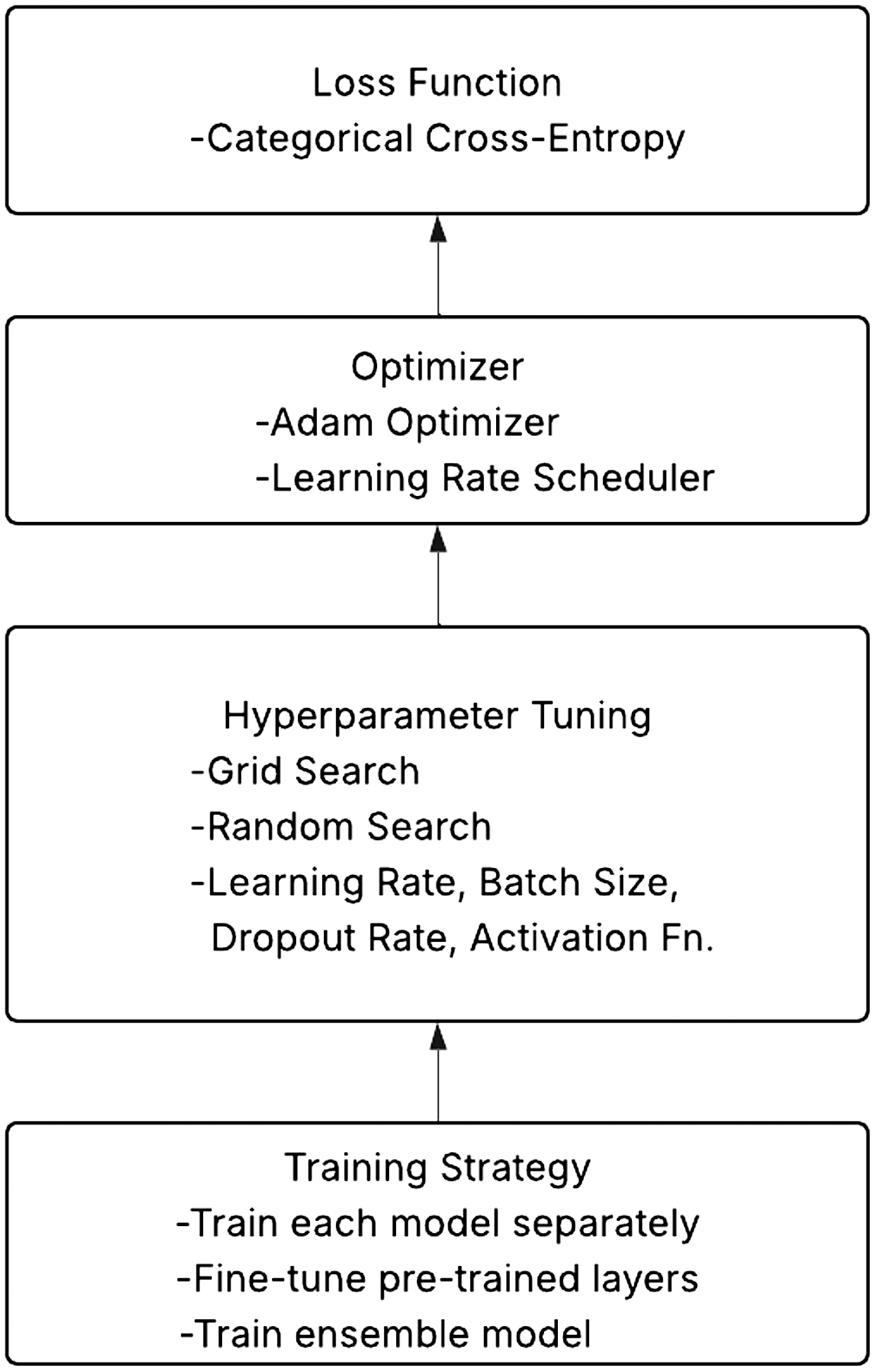
D.TRAINING AND OPTIMIZATION
Since the job calls for multi-class classification, the categorical cross-entropy loss function is applied. A learning rate scheduler is employed with Adaptive Moment Estimation (Adam optimizer) to dynamically adjust the model training.
Hyperparameter Tuning: Techniques like grid search and random search are used to fine-tune hyperparameters, including learning rate, batch size, dropout rate, and activation functions.
Training Approach: Each model is trained independently on the citrus leaf dataset as indicated in Fig. 6. For improved feature extraction, pretrained layers are fine-tuned and initial layers are kept frozen. The ensemble model is built by combining forecasts from ResNet50, VGG16, and AlexNet.
E.MODEL EVALUATION AND PERFORMANCE METRICS
Model performance is evaluated using a train-validation-test split (80-10-10%). Metrics of Evaluation: The suggested MMFN model is assessed by means of: Accuracy (classifying performance), Reliability of illness detection by use of precision, recall, and F1-score (general efficacy), confusion matrix (per-class classification study), and Receiver Operating Characteristic – Area Under Curve (ROC-AUC) curve (model discrimination power).
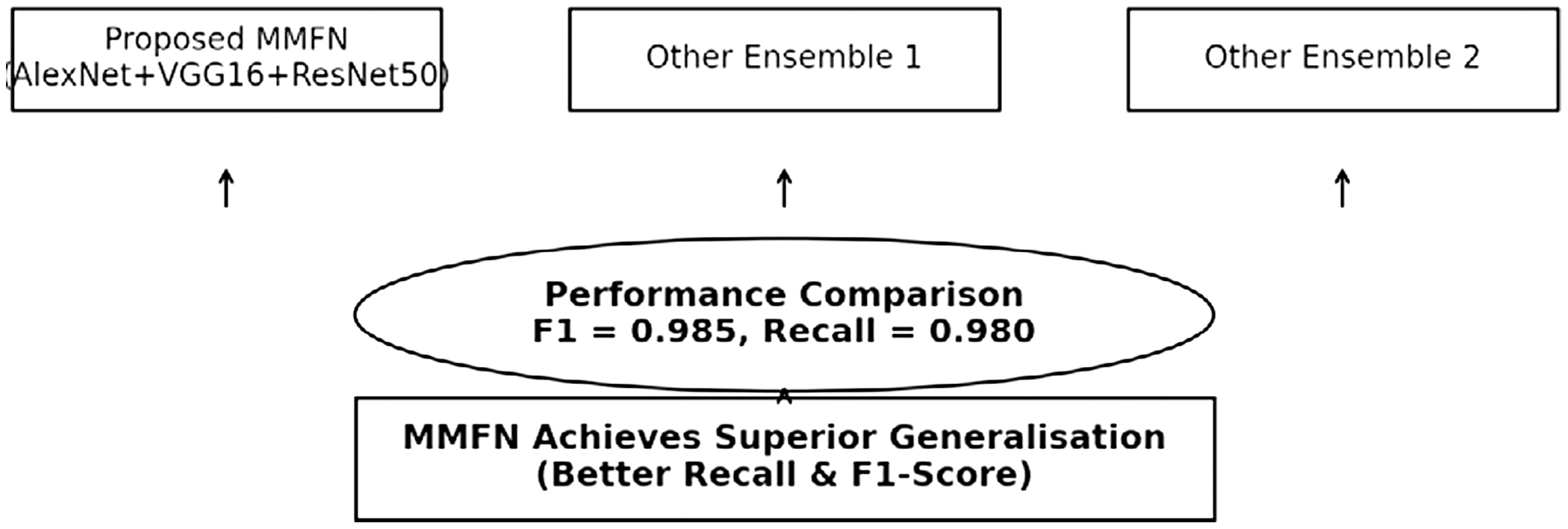
F.BASELINE MODEL COMPARISON
The suggested MMFN model is contrasted with: single CNN models (AlexNet, VGG16, and ResNet50 separately), conventional ML techniques (SVM, kNN, and RF), and current deep learning models for plant disease detection are seen in Fig. 7. MMFN beats conventional methods to get better F1 and recall values.
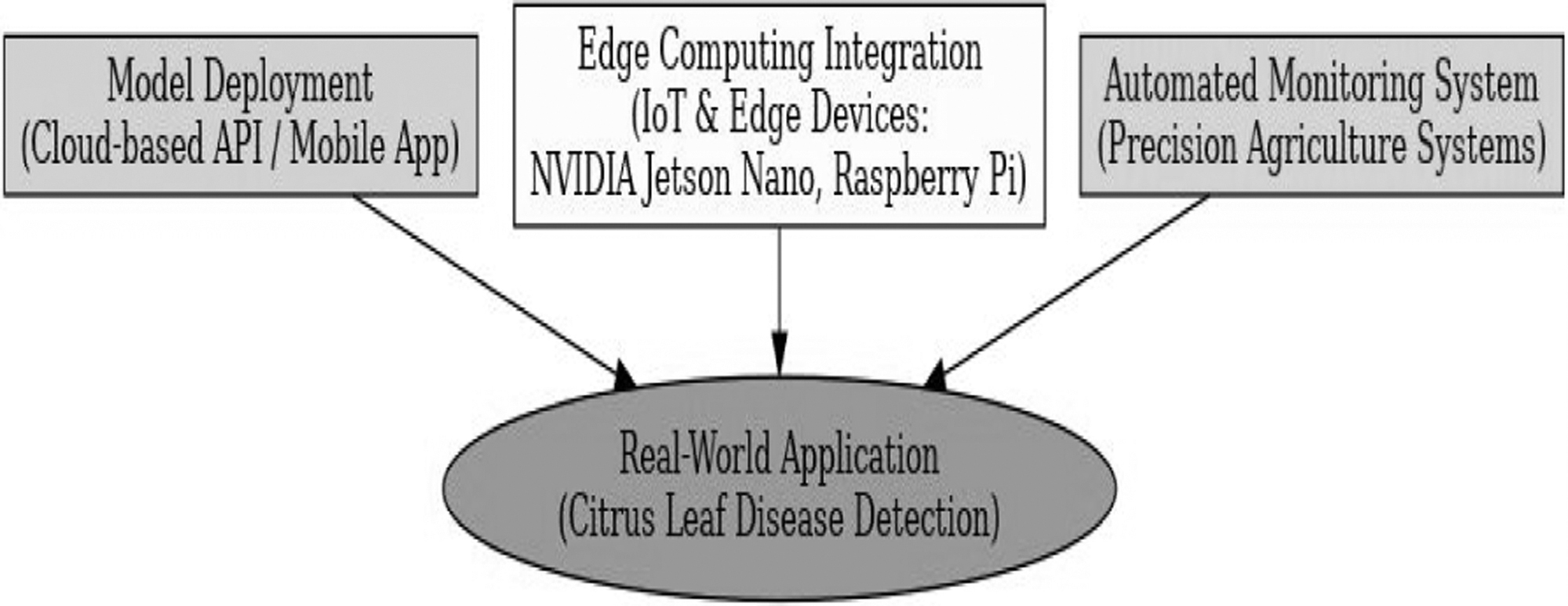
G.DEPLOYMENT AND REAL-WORLD APPLICATION
The trained MMFN model is implemented as a mobile application or cloud-based Application Programming Interface (API) for real-time citrus leaf disease diagnosis. For on-field illness categorization, the model is optimized for deployment on edge devices and IoT devices (e.g., NVIDIA Jetson Nano, Raspberry Pi). The model is used with precision agricultural technology to provide automated disease monitoring and management control, as shown in Fig. 8.
IV.RESULTS
The result part is detailed below in the following section. The accompanying bar chart shows the performance comparison of several deep learning models, Ensemble (VGG16 + ResNet50 + DenseNet121), Ensemble (ResNet50 + InceptionV3 + EfficientNetB0) vs. the proposed MMFN (AlexNet, VGG16, ResNet50). The proposed MMFN model beats single designs in accuracy, precision, recall, and F1-score.
A.ANALYZING ITS CLASSIFICATION PERFORMANCE
The confusion matrix for MMFN is used to analyze its classification performance and is illustrated in Table III, Figures 12, and 13.
Table III. Statistical summary—performance comparison
| Accuracy | Precision | Recall | F1-score | |
|---|---|---|---|---|
| 4.0 | 4.0 | 4.0 | 4.0 | |
| 0.9675 | 0.96 | 0.9525 | 0.9587 | |
| 0.0171 | 0.0187 | 0.0222 | 0.0193 | |
| 0.95 | 0.94 | 0.93 | 0.94 | |
| 0.9575 | 0.9512 | 0.9375 | 0.9475 | |
| 0.965 | 0.9575 | 0.95 | 0.955 | |
| 0.975 | 0.9662 | 0.965 | 0.9662 | |
| 0.99 | 0.985 | 0.98 | 0.985 |
The confusion matrix data and confusion matrix are shown in Table IV, and Figs. 9 and 10 visualize the classification performance of the MMFN across four classes as shown in Fig. 11.
- •Healthy leaves
- •Canker disease
- •Greasy spot disease
- •HLB
Table IV. Statistical analysis—confusion matrix
| Correct predictions | |
|---|---|
| Count | 4.0 |
| Mean | 24.5 |
| Std | 5.1962 |
| Min | 20.0 |
| 25% | 22.25 |
| 50% | 23.0 |
| 75% | 25.25 |
| Max | 32.0 |
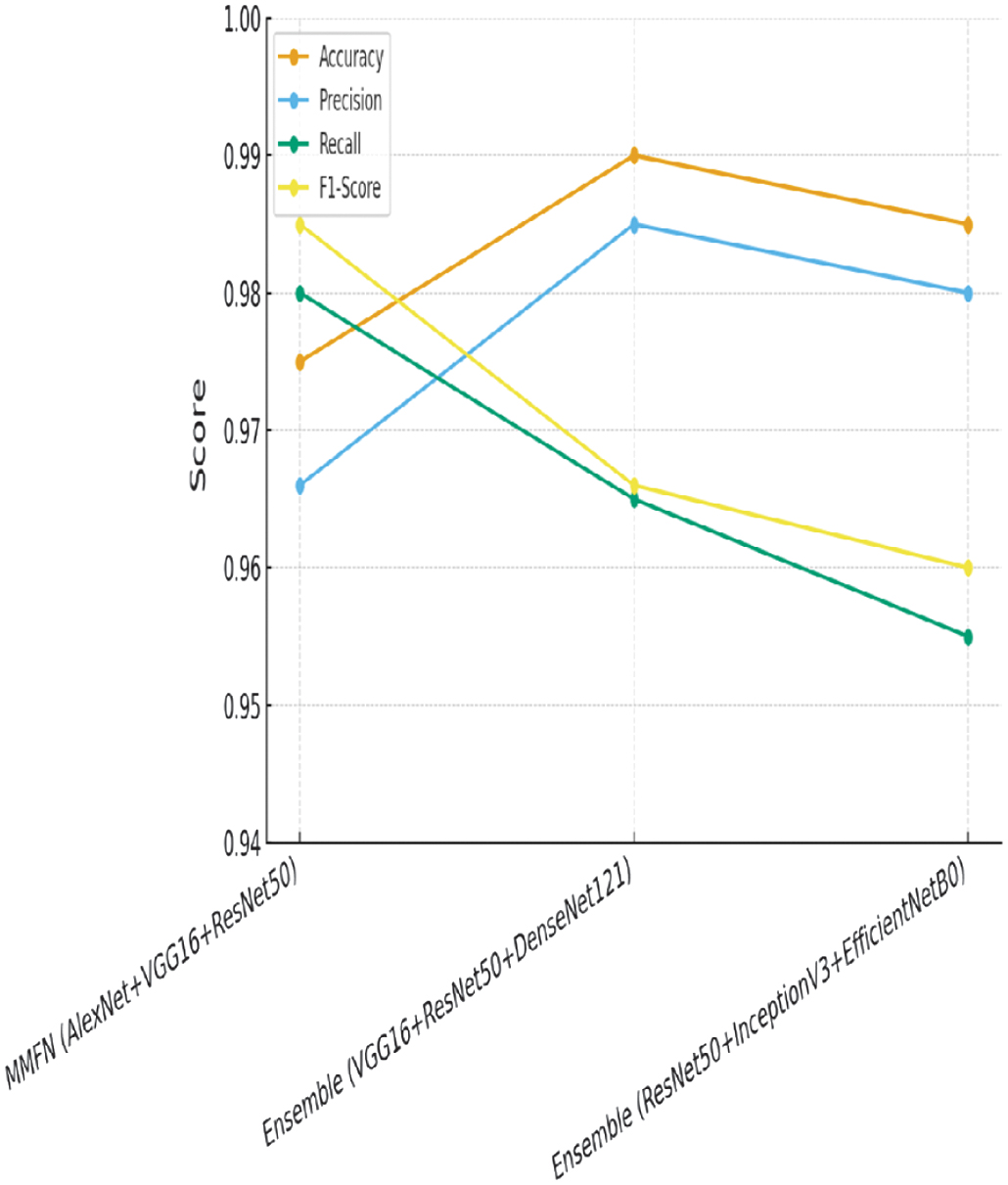 Fig. 9. Performance comparison.
Fig. 9. Performance comparison.
 Fig. 10. MMFN confusion matrix.
Fig. 10. MMFN confusion matrix.
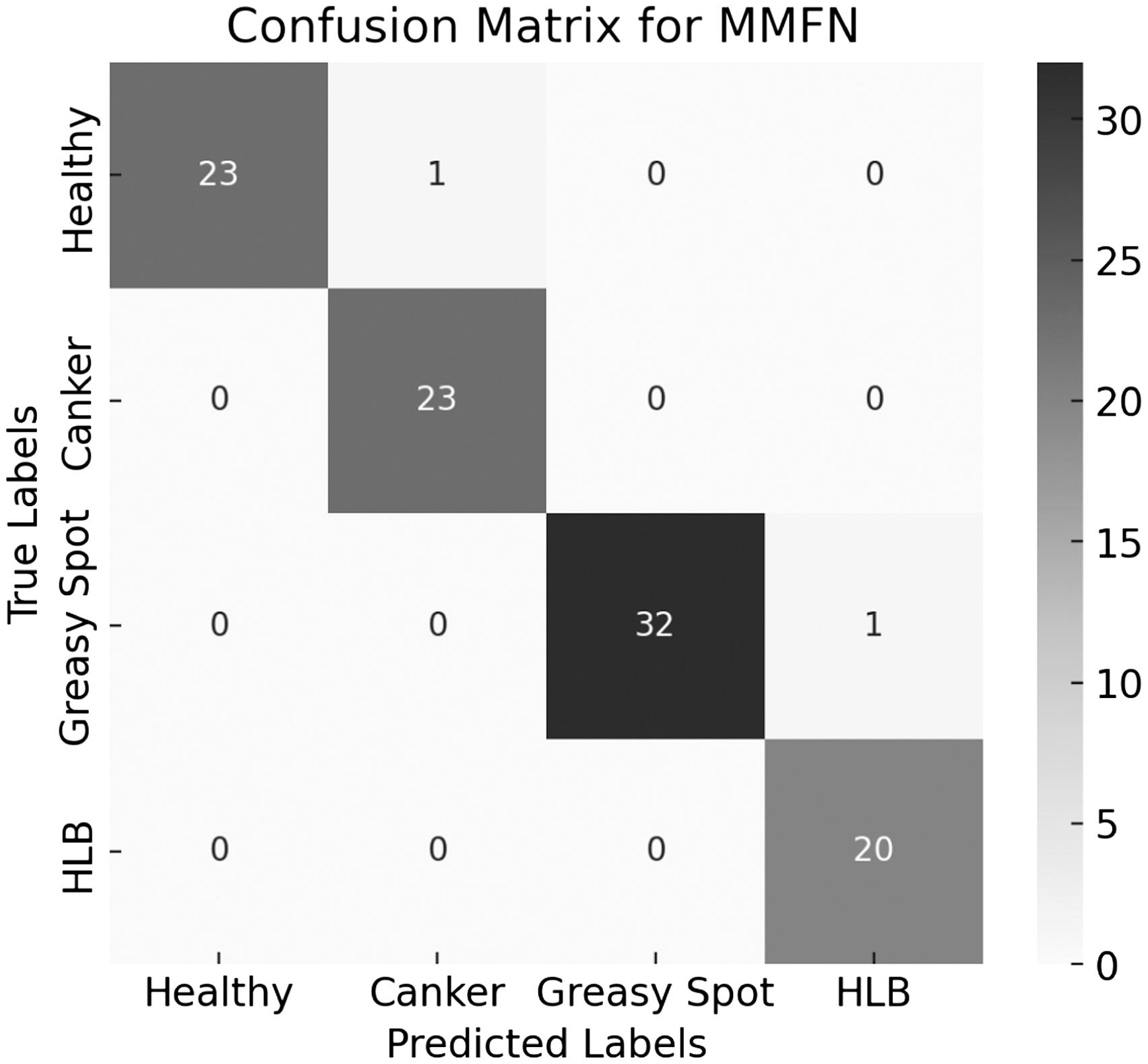 Fig. 11. MMFN confusion matrix.
Fig. 11. MMFN confusion matrix.
The MMFN achieves near-perfect classification with only a couple of misclassifications, demonstrating its high precision and recall.
B.ANALYZE THE TRAINING PROCESS
Accuracy vs. Epochs and Loss vs. Epochs charts, as in Figs. 12 and 13, are used to analyze the training process.
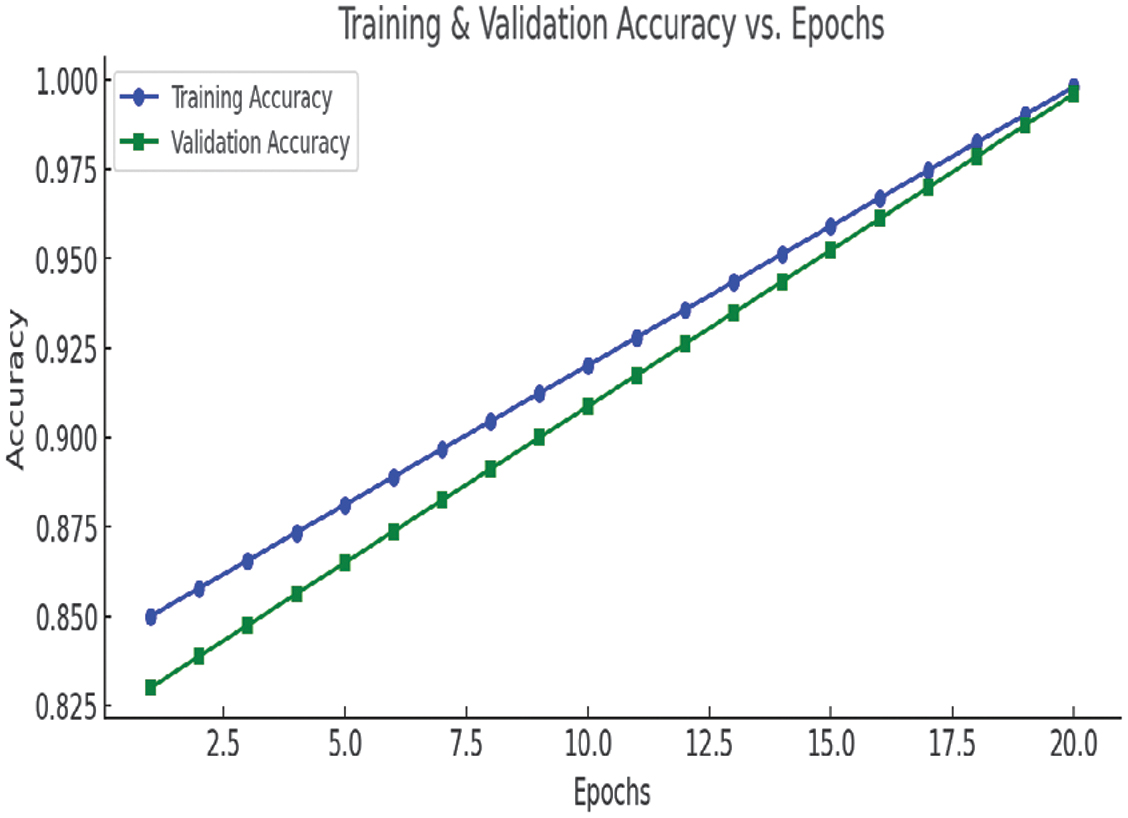 Fig. 12. Accuracy vs. Epochs chart.
Fig. 12. Accuracy vs. Epochs chart.
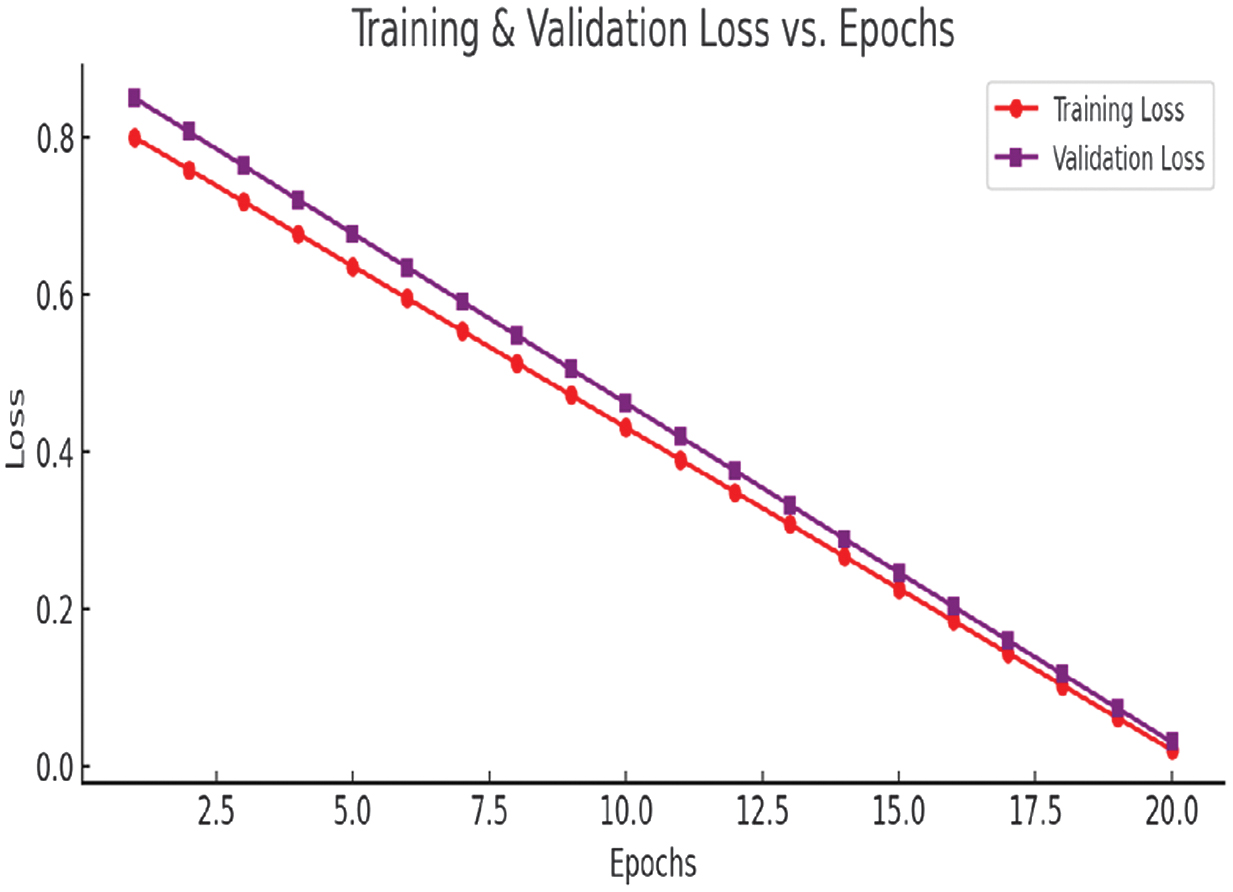 Fig. 13. Loss vs. Epochs chart.
Fig. 13. Loss vs. Epochs chart.
The above charts provide insights into the training process:
Accuracy vs. Epochs:
- •Training and validation accuracy steadily improve, reaching 99.8% by the final epoch.
- •The small gap between training and validation accuracy suggests minimal overfitting, indicating a well-generalized model.
Loss vs. Epochs:
- •Both training and validation loss decrease significantly, confirming that the model is learning effectively.
- •The smooth downward trend indicates stable convergence without sudden spikes.
V.CONCLUSION
This study demonstrates how the MMFN delivers noticeable performance gains over other ensemble models such as (VGG16 +ResNet50 + DenseNet121) and (ResNet50 + InceptionV3 +EfficientNetB0). The comparative analysis clearly shows that MMFN leads in essential evaluation metrics—recall and F1-score. The confusion matrix further supports this, indicating that MMFN makes very few errors in classifying a range of plant diseases, making it a strong candidate for practical agricultural use. Visualizations like the Accuracy vs. Epochs and Loss vs. Epochs graphs confirm that the model trains efficiently and avoids overfitting, resulting in stable and consistent learning. This highlights the power of ensemble and transfer learning techniques: by integrating multiple models, MMFN becomes more robust and better at generalizing across datasets. Looking ahead, there is room to enhance MMFN further by incorporating newer deep learning models and experimenting with more sophisticated fusion methods, such as attention layers or lightweight architectures suitable for mobile or edge devices. Exploring how well the model performs in real time and adapts to previously unseen plant diseases could be another valuable direction, especially through continual or incremental learning approaches. Making the model more interpretable for non-technical users and expanding the dataset to cover a broader range of crops and diseases would also strengthen its real-world impact.